Pectin methylesterase gene AtPMEPCRA contributes to physiological adaptation to simulated and spaceflight microgravity in Arabidopsis
- PMID: 35602950
- PMCID: PMC9118689
- DOI: 10.1016/j.isci.2022.104331
Pectin methylesterase gene AtPMEPCRA contributes to physiological adaptation to simulated and spaceflight microgravity in Arabidopsis
Abstract
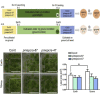
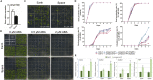
Pectin is biosynthesized in a highly methylated form and is partially de-methylated by pectin methylesterase (PME) activity. Plant PMEs play a critical role in cell wall remodeling in many physiological processes. Here, we studied Arabidopsis seedlings, which had been exposed to simulated or actual microgravity. Simulated microgravity inhibited total PME activity in Arabidopsis seedlings. We identified that AtPMEPCRA expression played a major role in the microgravity-induced inhibition of PME activity. atpmepcra mutants did not exhibit the enlarged leaf area of Arabidopsis seedlings observed under spaceflight microgravity. The downregulation of AtPMEPCRA expression in response to microgravity was due, in part, to changes in methylation patterns. The sexual offspring of the plants grown during spaceflight retained the methylation changes at AtPMEPCRA locus for one generation and thus contribute to the physiological adaptation to microgravity among F1 offspring seed generation. We conclude that AtPMEPCRA contributes to the spaceflight-induced transgenerational responses in Arabidopsis.
Keywords: Biological sciences; Plant biology; Plant physiology; Space sciences.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

