Proteome Profiling Identifies Serum Biomarkers in Rheumatoid Arthritis
- PMID: 35603148
- PMCID: PMC9120366
- DOI: 10.3389/fimmu.2022.865425
Proteome Profiling Identifies Serum Biomarkers in Rheumatoid Arthritis
Abstract
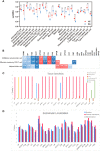
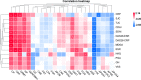
Rheumatoid arthritis (RA) causes serious disability and productivity loss, and there is an urgent need for appropriate biomarkers for diagnosis, treatment assessment, and prognosis evaluation. To identify serum markers of RA, we performed mass spectrometry (MS)-based proteomics, and we obtained 24 important markers in normal and RA patient samples using a random forest machine learning model and 11 protein-protein interaction (PPI) network topological analysis methods. Markers were reanalyzed using additional proteomics datasets, immune infiltration status, tissue specificity, subcellular localization, correlation analysis with disease activity-based diagnostic indications, and diagnostic receiver-operating characteristic analysis. We discovered that ORM1 in serum is significantly differentially expressed in normal and RA patient samples, which is positively correlated with disease activity, and is closely related to CD56dim natural killer cell, effector memory CD8+T cell, and natural killer cell in the pathological mechanism, which can be better utilized for future research on RA. This study supplies a comprehensive strategy for discovering potential serum biomarkers of RA and provides a different perspective for comprehending the pathological mechanism of RA, identifying potential therapeutic targets, and disease management.
Keywords: ORM1; biomarker; proteomics; rheumatoid arthritis; serum.
Copyright © 2022 Hu, Dai, Xu, Zhao, Xu, Li, Yu, Zhang, Deng, Liu, Zhang, Huang, Wu and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Machine learning and weighted gene co-expression network analysis identify a three-gene signature to diagnose rheumatoid arthritis.Front Immunol. 2024 Apr 22;15:1387311. doi: 10.3389/fimmu.2024.1387311. eCollection 2024. Front Immunol. 2024. PMID: 38711508 Free PMC article.
-
Identification of diagnostic genes and drug prediction in metabolic syndrome-associated rheumatoid arthritis by integrated bioinformatics analysis, machine learning, and molecular docking.Front Immunol. 2024 Jul 29;15:1431452. doi: 10.3389/fimmu.2024.1431452. eCollection 2024. Front Immunol. 2024. PMID: 39139563 Free PMC article.
-
Identification of CD8+ T cell-related biomarkers and immune infiltration characteristic of rheumatoid arthritis.Aging (Albany NY). 2024 Jan 16;16(2):1399-1413. doi: 10.18632/aging.205435. Epub 2024 Jan 16. Aging (Albany NY). 2024. PMID: 38231477 Free PMC article.
-
Defining the proteomic landscape of rheumatoid arthritis: progress and prospective clinical applications.Expert Rev Proteomics. 2017 May;14(5):431-444. doi: 10.1080/14789450.2017.1321481. Epub 2017 May 2. Expert Rev Proteomics. 2017. PMID: 28425787 Review.
-
Biomarkers in Rheumatoid Arthritis, what is new?J Med Life. 2016 Apr-Jun;9(2):144-8. J Med Life. 2016. PMID: 27453744 Free PMC article. Review.
Cited by
-
Proteomic Profiling Reveals Novel Molecular Insights into Dysregulated Proteins in Established Cases of Rheumatoid Arthritis.Proteomes. 2025 Jul 4;13(3):32. doi: 10.3390/proteomes13030032. Proteomes. 2025. PMID: 40700276 Free PMC article.
-
A longitudinal cohort study uncovers plasma protein biomarkers predating clinical onset and treatment response of rheumatoid arthritis.Nat Commun. 2025 Jul 21;16(1):6692. doi: 10.1038/s41467-025-62032-1. Nat Commun. 2025. PMID: 40691443 Free PMC article.
-
Serum Proteomic Analysis Revealed Biomarkers for Eosinophilic Chronic Rhinosinusitis with Nasal Polyps Pathophysiology.J Inflamm Res. 2024 Feb 7;17:805-821. doi: 10.2147/JIR.S444280. eCollection 2024. J Inflamm Res. 2024. PMID: 38344304 Free PMC article.
-
Identification of Plasma Biomarkers from Rheumatoid Arthritis Patients Using an Optimized Sequential Window Acquisition of All THeoretical Mass Spectra (SWATH) Proteomics Workflow.Proteomes. 2023 Oct 16;11(4):32. doi: 10.3390/proteomes11040032. Proteomes. 2023. PMID: 37873874 Free PMC article.
-
Serum Biomarkers of Vascular Involvement in Childhood Uveitis.Transl Vis Sci Technol. 2024 Apr 2;13(4):9. doi: 10.1167/tvst.13.4.9. Transl Vis Sci Technol. 2024. PMID: 38573655 Free PMC article.
References
-
- Glick EN. Immunopathogenesis of Rheumatoid Arthritis. J R Soc Med (1980) 73:80. doi: 10.1177/014107688007300129 - DOI
-
- Wells G, Li T, Maxwell L, Maclean R, Tugwell P. Responsiveness of Patient Reported Outcomes Including Fatigue, Sleep Quality, Activity Limitation, and Quality of Life Following Treatment With Abatacept for Rheumatoid Arthritis. Ann Rheumat Dis (2008) 67:260–5. doi: 10.1136/ard.2007.069690 - DOI - PubMed
-
- Kroot EJJA, Jong B, Leeuwen M, Swinkels H, Hoogen F.H.J.V.D., Hof MVT, et al. The Prognostic Value of Anti-Cyclic Citrullinated Peptide Antibody in Patients With Recent-Onset Rheumatoid Arthritis. Arthritis Rheumatol (2000) 43:1831–5. doi: 10.1002/1529-0131(200008)43:8<1831::AID-ANR19>3.0.CO;2-6 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

