Estimating the COVID-19 infection fatality ratio accounting for seroreversion using statistical modelling
- PMID: 35603270
- PMCID: PMC9120146
- DOI: 10.1038/s43856-022-00106-7
Estimating the COVID-19 infection fatality ratio accounting for seroreversion using statistical modelling
Abstract
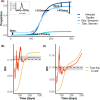
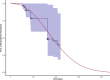
Background: The infection fatality ratio (IFR) is a key statistic for estimating the burden of coronavirus disease 2019 (COVID-19) and has been continuously debated throughout the COVID-19 pandemic. The age-specific IFR can be quantified using antibody surveys to estimate total infections, but requires consideration of delay-distributions from time from infection to seroconversion, time to death, and time to seroreversion (i.e. antibody waning) alongside serologic test sensitivity and specificity. Previous IFR estimates have not fully propagated uncertainty or accounted for these potential biases, particularly seroreversion.
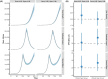
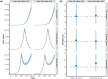
Methods: We built a Bayesian statistical model that incorporates these factors and applied this model to simulated data and 10 serologic studies from different countries.
Results: We demonstrate that seroreversion becomes a crucial factor as time accrues but is less important during first-wave, short-term dynamics. We additionally show that disaggregating surveys by regions with higher versus lower disease burden can inform serologic test specificity estimates. The overall IFR in each setting was estimated at 0.49-2.53%.
Conclusion: We developed a robust statistical framework to account for full uncertainties in the parameters determining IFR. We provide code for others to apply these methods to further datasets and future epidemics.
Keywords: Computational biology and bioinformatics; Respiratory tract diseases.
© The Author(s) 2022.
Conflict of interest statement
Competing interestsPGTW is an Editorial Board Member for Communications Medicine, but was not involved in the editorial review or peer review, nor in the decision to publish this article. The other authors have no competing interests to declare.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

