Salmonella Induces the cGAS-STING-Dependent Type I Interferon Response in Murine Macrophages by Triggering mtDNA Release
- PMID: 35604097
- PMCID: PMC9239183
- DOI: 10.1128/mbio.03632-21
Salmonella Induces the cGAS-STING-Dependent Type I Interferon Response in Murine Macrophages by Triggering mtDNA Release
Abstract
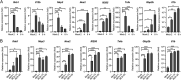
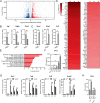
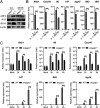
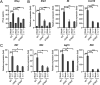
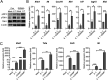
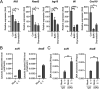
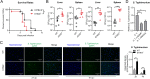
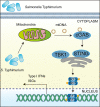
Salmonella enterica serovar Typhimurium (S. Typhimurium) elicited strong innate immune responses in macrophages. To activate innate immunity, pattern recognition receptors (PRRs) in host cells can recognize highly conserved pathogen-associated molecular patterns (PAMPs). Here, we showed that S. Typhimurium induced a robust type I interferon (IFN) response in murine macrophages. Exposure of macrophages to S. Typhimurium activated a Toll-like receptor 4 (TLR4)-dependent type I IFN response. Next, we showed that type I IFN and IFN-stimulated genes (ISGs) were elicited in a TBK1-IFN-dependent manner. Furthermore, cytosolic DNA sensor cyclic GMP-AMP synthase (cGAS) and immune adaptor protein stimulator of interferon genes (STING) were also required for the induction of type I IFN response during infection. Intriguingly, S. Typhimurium infection triggered mitochondrial DNA (mtDNA) release into the cytosol to activate the type I IFN response. In addition, we also showed that bacterial DNA was enriched in cGAS during infection, which may contribute to cGAS activation. Finally, we showed that cGAS and STING deficient mice and cells were more susceptible to S. Typhimurium infection, signifying the critical role of the cGAS-STING pathway in host defense against S. Typhimurium infection. In conclusion, in addition to TLR4-dependent innate immune response, we demonstrated that S. Typhimurium induced the type I IFN response in a cGAS-STING-dependent manner and the S. Typhimurium-induced mtDNA release was important for the induction of type I IFN. This study elucidated a new mechanism by which bacterial pathogen activated the cGAS-STING pathway and also characterized the important role of cGAS-STING during S. Typhimurium infection. IMPORTANCE As one of the most common foodborne transmitted zoonotic pathogens, S. Typhimurium infection causes diarrheal disease in humans and animals. S. Typhimurium infection has been implicated as an inducer for the type I interferon (IFN) response in macrophages, but the mechanisms are not fully understood. In this study, we reported that in addition to TLR4-dependent response, the cytosolic surveillance pathway (CSP) cGAS-STING is also required for the activation of type I IFN response during S. Typhimurium infection. We further showed that the infection of S. Typhimurium triggered mtDNA release into the cytosol, which induces the type I IFN response. In addition, physical interactions between cGAS and S. Typhimurium DNA have been identified in the context of infection. Importantly, we also provided convincing in vivo and in vitro evidence that the cGAS-STING pathway was potently implicated in the host defense against S. Typhimurium infection. Together, we uncovered a mechanism by which type I IFN response is elicited during S. Typhimurium infection in murine macrophages in an mtDNA-cGAS-STING-dependent manner.
Keywords: STING; Salmonella; cGAS; interferon; mtDNA; type I interferon.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Royle MC, Totemeyer S, Alldridge LC, Maskell DJ, Bryant CE. 2003. Stimulation of Toll-like receptor 4 by lipopolysaccharide during cellular invasion by live Salmonella typhimurium is a critical but not exclusive event leading to macrophage responses. J Immunol 170:5445–5454. doi: 10.4049/jimmunol.170.11.5445. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

