A novel Cbx1, PurB, and Sp3 complex mediates long-term silencing of tissue- and lineage-specific genes
- PMID: 35605661
- PMCID: PMC9190063
- DOI: 10.1016/j.jbc.2022.102053
A novel Cbx1, PurB, and Sp3 complex mediates long-term silencing of tissue- and lineage-specific genes
Abstract
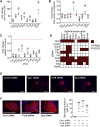
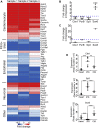
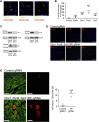
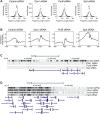
miRNA-based cellular fate reprogramming offers an opportunity to investigate the mechanisms of long-term gene silencing. To further understand how genes are silenced in a tissue-specific manner, we leveraged our miRNA-based method of reprogramming fibroblasts into cardiomyocytes. Through screening approaches, we identified three proteins that were downregulated during reprogramming of fibroblasts into cardiomyocytes: heterochromatin protein Cbx1, transcriptional activator protein PurB, and transcription factor Sp3. We show that knockdown of Cbx1, PurB, and Sp3 was sufficient to induce cardiomyocyte gene expression in fibroblasts. Similarly, gene editing to ablate Cbx1, PurB, and Sp3 expression induced fibroblasts to convert into cardiomyocytes in vivo. Furthermore, high-throughput DNA sequencing and coimmunoprecipitation experiments indicated that Cbx1, PurB, and Sp3 also bound together as a complex and were necessary to localize nucleosomes to cardiomyocyte genes on the chromosome. Finally, we found that the expression of these genes led to nucleosome modification via H3K27me3 (trimethylated histone-H3 lysine-27) deposition through an interaction with the polycomb repressive PRC2 complex. In summary, we conclude that Cbx1, PurB, and Sp3 control cell fate by actively repressing lineage-specific genes.
Keywords: cardiac development; cardiac muscle; cardiomyocyte; fibroblast; regeneration; reprogramming; transcription repressor.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest V. J. D. and C. P. H. are cofounders of Recardia Therapeutics. This company is focused on developing miRNAs that reprogram fibroblasts into cardiomyocytes. All other authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Sadahiro T., Ieda M. In vivo reprogramming as a new approach to cardiac regenerative therapy. Semin. Cell Dev. Biol. 2022;122:21–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

