p53 wild-type colorectal cancer cells that express a fetal gene signature are associated with metastasis and poor prognosis
- PMID: 35606354
- PMCID: PMC9126967
- DOI: 10.1038/s41467-022-30382-9
p53 wild-type colorectal cancer cells that express a fetal gene signature are associated with metastasis and poor prognosis
Abstract
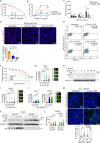
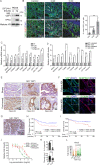
Current therapy against colorectal cancer (CRC) is based on DNA-damaging agents that remain ineffective in a proportion of patients. Whether and how non-curative DNA damage-based treatment affects tumor cell behavior and patient outcome is primarily unstudied. Using CRC patient-derived organoids (PDO)s, we show that sublethal doses of chemotherapy (CT) does not select previously resistant tumor populations but induces a quiescent state specifically to TP53 wildtype (WT) cancer cells, which is linked to the acquisition of a YAP1-dependent fetal phenotype. Cells displaying this phenotype exhibit high tumor-initiating and metastatic activity. Nuclear YAP1 and fetal traits are present in a proportion of tumors at diagnosis and predict poor prognosis in patients carrying TP53 WT CRC tumors. We provide data indicating the higher efficacy of CT together with YAP1 inhibitors for eradication of therapy resistant TP53 WT cancer cells. Together these results identify fetal conversion as a useful biomarker for patient prognosis and therapy prescription.
© 2022. The Author(s).
Conflict of interest statement
Laura Solé, Teresa Lobo-Jarne, Marta Guix, Beatriz Bellosillo, Mar Iglesias, Anna Bigas, and Lluís Espinosa have a pending patent application entitled Genomic predictor of outcome in cancer with number PCT/EP2022/058503, related to the fetal signature as a prognostic biomarker in colorectal cancer. The authors have no additional financial interests and the remaining authors declare no competing interests.
Figures




References
-
- Brenner, H., Kloor, M. & Pox, C. P. Colorectal cancer. Lancet10.1016/S0140-6736(13)61649-9 (2014). - PubMed
-
- Batlle, E. & Clevers, H. Cancer stem cells revisited. Nat. Med.10.1038/nm.4409 (2017). - PubMed
-
- Galmarini, F. C., Galmarini, C. M., Sarchi, M. I., Abulafia, J. & Galmarini, D. Heterogeneous distribution of tumor blood supply affects the response to chemotherapy in patients with head and neck cancer. Microcirculation7, 405–410 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

