Epithelial de-differentiation triggered by co-ordinate epigenetic inactivation of the EHF and CDX1 transcription factors drives colorectal cancer progression
- PMID: 35606410
- PMCID: PMC9613692
- DOI: 10.1038/s41418-022-01016-w
Epithelial de-differentiation triggered by co-ordinate epigenetic inactivation of the EHF and CDX1 transcription factors drives colorectal cancer progression
Abstract
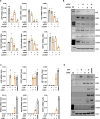
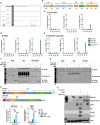
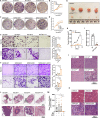
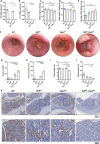
Colorectal cancers (CRCs) often display histological features indicative of aberrant differentiation but the molecular underpinnings of this trait and whether it directly drives disease progression is unclear. Here, we identify co-ordinate epigenetic inactivation of two epithelial-specific transcription factors, EHF and CDX1, as a mechanism driving differentiation loss in CRCs. Re-expression of EHF and CDX1 in poorly-differentiated CRC cells induced extensive chromatin remodelling, transcriptional re-programming, and differentiation along the enterocytic lineage, leading to reduced growth and metastasis. Strikingly, EHF and CDX1 were also able to reprogramme non-colonic epithelial cells to express colonic differentiation markers. By contrast, inactivation of EHF and CDX1 in well-differentiated CRC cells triggered tumour de-differentiation. Mechanistically, we demonstrate that EHF physically interacts with CDX1 via its PNT domain, and that these transcription factors co-operatively drive transcription of the colonic differentiation marker, VIL1. Compound genetic deletion of Ehf and Cdx1 in the mouse colon disrupted normal colonic differentiation and significantly enhanced colorectal tumour progression. These findings thus reveal a novel mechanism driving epithelial de-differentiation and tumour progression in CRC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Zong L, Abe M, Ji J, Zhu W-G, Yu D. Tracking the correlation between CpG island methylator phenotype and other molecular features and clinicopathological features in human colorectal cancers: a systematic review and meta-analysis. Clin Transl Gastroenterol. 2016;7:e151. doi: 10.1038/ctg.2016.14. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

