Single-domain near-infrared protein provides a scaffold for antigen-dependent fluorescent nanobodies
- PMID: 35606446
- PMCID: PMC9189029
- DOI: 10.1038/s41592-022-01467-6
Single-domain near-infrared protein provides a scaffold for antigen-dependent fluorescent nanobodies
Abstract
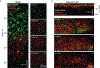
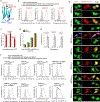
Small near-infrared (NIR) fluorescent proteins (FPs) are much needed as protein tags for imaging applications. We developed a 17 kDa NIR FP, called miRFP670nano3, which brightly fluoresces in mammalian cells and enables deep-brain imaging. By exploring miRFP670nano3 as an internal tag, we engineered 32 kDa NIR fluorescent nanobodies, termed NIR-Fbs, whose stability and fluorescence strongly depend on the presence of specific intracellular antigens. NIR-Fbs allowed background-free visualization of endogenous proteins, detection of viral antigens, labeling of cells expressing target molecules and identification of double-positive cell populations with bispecific NIR-Fbs against two antigens. Applying NIR-Fbs as destabilizing fusion partners, we developed molecular tools for directed degradation of targeted proteins, controllable protein expression and modulation of enzymatic activities. Altogether, NIR-Fbs enable the detection and manipulation of a variety of cellular processes based on the intracellular protein profile.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures




Comment in
-
New near-infrared fluorescent probes and tools.Nat Methods. 2022 Jun;19(6):654-655. doi: 10.1038/s41592-022-01425-2. Nat Methods. 2022. PMID: 35606447 No abstract available.
References
-
- Chernov KG, Redchuk TA, Omelina ES & Verkhusha VV Near-Infrared Fluorescent Proteins, Biosensors, and Optogenetic Tools Engineered from Phytochromes. Chem Rev 117, 6423–6446 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

