Molecular characteristics and genetic evolutionary analyses of circulating parvoviruses derived from cats in Beijing
- PMID: 35606875
- PMCID: PMC9125828
- DOI: 10.1186/s12917-022-03281-w
Molecular characteristics and genetic evolutionary analyses of circulating parvoviruses derived from cats in Beijing
Abstract
Background: Feline parvovirus (FPV) is a member of the family Parvoviridae, which is a major enteric pathogen of cats worldwide. This study aimed to investigate the prevalence of feline parvovirus in Beijing of China and analyze the genetic features of detected viruses.
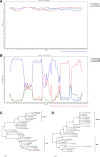
Results: In this study, a total of 60 (8.5%) parvovirus-positive samples were detected from 702 cat fecal samples using parvovirus-specific PCR. The complete VP2 genes were amplified from all these samples. Among them, 55 (91.7%) sequences were characterized as FPV, and the other five (8.3%) were typed as canine parvovirus type 2 (CPV-2) variants, comprised of four CPV-2c and a new CPV-2b strain. In order to investigate the origin of CPV-2 variants in cats, we amplified full-length VP2 genes from seven fecal samples of dogs infected with CPV-2, which were further classified as CPV-2c. The sequences of new CPV-2b/MT270586 and CPV-2c/MT270587 detected from feline samples shared 100% identity with previous canine isolates KT156833 and MF467242 respectively, suggesting the CPV-2 variants circulating in cats might be derived from dogs. Sequence analysis indicated new mutations, Ala91Ser and Ser192Phe, in the FPV sequences, while obtained CPV-2c carried mutations reported in Asian CPV variants, showing they share a common evolutionary pattern with the Asian 2c strains. Interestingly, the FPV sequence (MT270571), displaying four CPV-specific residues, was found to be a putative recombinant sequence between CPV-2c and FPV. Phylogenetic analysis of the VP2 gene showed that amino acid and nucleotide mutations promoted the evolution of FPV and CPV lineages.
Conclusions: Our findings will be helpful to further understand the circulation and evolution of feline and canine parvovirus in Beijing.
Keywords: Canine parvovirus (CPV); Evolution; Feline parvovirus (FPV); Recombination; VP2 gene.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Niu J, Yi S, Hu G, Guo Y, Zhang S, Dong H, Zhao Y, Wang K. Prevalence and molecular characterization of parvovirus in domestic kittens from Northeast China during 2016–2017. Jpn J Vet Res. 2018;66(3):145–155.
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

