Serum proteome alterations during conventional and extracorporeal resuscitation in pigs
- PMID: 35606879
- PMCID: PMC9125930
- DOI: 10.1186/s12967-022-03441-4
Serum proteome alterations during conventional and extracorporeal resuscitation in pigs
Abstract
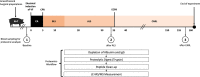
Background: Only a small number of patients survive an out-of-hospital cardiac arrest (CA) and can be discharged from hospital alive with a large percentage of these patients retaining neurological impairments. In recent years, extracorporeal cardiopulmonary resuscitation (ECPR) has emerged as a beneficial strategy to optimize cardiac arrest treatment. However, ECPR is still associated with various complications. To reduce these problems, a profound understanding of the underlying mechanisms is required. This study aims to investigate the effects of CA, conventional cardiopulmonary resuscitation (CPR) and ECPR using a whole-body reperfusion protocol (controlled and automated reperfusion of the whole body-CARL) on the serum proteome profiles in a pig model of refractory CA.
Methods: N = 7 pigs underwent 5 min of untreated CA followed by 30 min CPR and 120 min perfusion with CARL. Blood samples for proteomic analysis were drawn at baseline, after CPR and at the end of the CARL period. Following albumin-depletion, proteomic analysis was performed using liquid chromatography-tandem mass spectrometry.
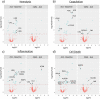
Results: N = 21 serum samples were measured resulting in the identification and quantification of 308-360 proteins per sample and 388 unique proteins in total. The three serum proteome profiles at the investigated time points clustered individually and segregated almost completely when considering a 90% confidence interval. Differential expression analysis showed significant abundance changes in 27 proteins between baseline and after CPR and in 9 proteins after CARL compared to CPR. Significant findings were further validated through a co-abundance cluster analysis corroborating the observed abundance changes.
Conclusions: The presented data highlight the impact of systemic ischemia and reperfusion on the entire serum proteome during resuscitation with a special focus on changes regarding haemolysis, coagulation, inflammation, and cell-death processes. Generally, the observed changes contribute to post-ischemic complications. Better understanding of the underlying mechanisms during CA and resuscitation may help to limit these complications and improve therapeutic options.
Keywords: Cardiopulmonary resuscitation; Extracorporeal cardiopulmonary resuscitation; Extracorporeal membrane oxygenation; Ischemia reperfusion injury; Proteome.
© 2022. The Author(s).
Conflict of interest statement
FB, CB and GT are shareholders in Resuscitec GmbH, Freiburg, Germany, which is a start-up company from the University Medical Centre Freiburg. JSP, SJB and CS are part-time employees of Resuscitec GmbH, Freiburg, Germany. The remaining authors have disclosed that they do not have any conflicts of interest.
Figures


References
-
- Guy A, Kawano T, Besserer F, Scheuermeyer F, Kanji HD, Christenson J, et al. The relationship between no-flow interval and survival with favourable neurological outcome in out-of-hospital cardiac arrest: implications for outcomes and ECPR eligibility. Resuscitation. 2020;155:219–225. doi: 10.1016/j.resuscitation.2020.06.009. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

