A prospective matched case-control study on the genomic epidemiology of colistin-resistant Enterobacterales from Dutch patients
- PMID: 35607432
- PMCID: PMC9122983
- DOI: 10.1038/s43856-022-00115-6
A prospective matched case-control study on the genomic epidemiology of colistin-resistant Enterobacterales from Dutch patients
Abstract
Background: Colistin is a last-resort treatment option for infections with multidrug-resistant Gram-negative bacteria. However, colistin resistance is increasing.
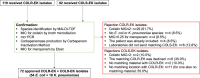
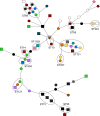
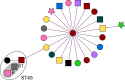
Methods: A six-month prospective matched case-control study was performed in which 22 Dutch laboratories with 32 associated hospitals participated. Laboratories were invited to send a maximum of five colistin-resistant Escherichia coli or Klebsiella pneumoniae (COLR-EK) isolates and five colistin-susceptible isolates (COLS-EK) to the reference laboratory, matched for patient location, material of origin and bacterial species. Epidemiological/clinical data were collected and included in the analysis. Characteristics of COLR-EK/COLS-EK isolates were compared using logistic regression with correction for variables used for matching. Forty-six ColR-EK/ColS-EK pairs were analysed by next-generation sequencing (NGS) for whole-genome multi-locus sequence typing and identification of resistance genes, including mcr genes. To identify chromosomal mutations potentially leading to colistin resistance, NGS reads were mapped against gene sequences of pmrAB, phoPQ, mgrB and crrB.
Results: In total, 72 COLR-EK/COLS-EK pairs (75% E. coli and 25% K. pneumoniae) were included. Twenty-one percent of COLR-EK patients had received colistin, in contrast to 3% of COLS-EK patients (OR > 2.9). Of COLR-EK isolates, five contained mcr-1 and two mcr-9. One isolate lost mcr-9 after repeated sub-culturing, but retained colistin resistance. Among 46 sequenced COLR-EK isolates, genetic diversity was large and 19 (41.3%) isolates had chromosomal mutations potentially associated with colistin resistance.
Conclusions: Colistin resistance is present but uncommon in the Netherlands and caused by the mcr gene in a minority of COLR-EK isolates. There is a need for surveillance of colistin resistance using appropriate susceptibility testing methods.
Keywords: Antimicrobial resistance; Epidemiology; Infectious-disease epidemiology.
© The Author(s) 2022.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures
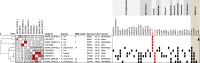
References
-
- World Health Organization. Central Asian and Eastern European Surveillance of Antimicrobial Resistance. Annual Report 2017. https://www.euro.who.int/en/health-topics/disease-prevention/antimicrobi... (2018).
-
- European Centre for Disease Prevention and Control. Solna: Antimicrobial resistance in the EU/EEA (EARS-Net). Annual Epidemiological Report 2019. https://www.ecdc.europa.eu/sites/default/files/documents/surveillance-an... (2020).
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

