Affinity Chromatography-Based Assays for the Screening of Potential Ligands Selective for G-Quadruplex Structures
- PMID: 35608081
- PMCID: PMC9127747
- DOI: 10.1002/open.202200090
Affinity Chromatography-Based Assays for the Screening of Potential Ligands Selective for G-Quadruplex Structures
Abstract
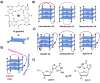
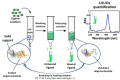
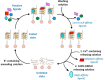
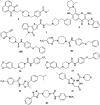
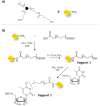
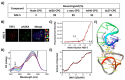
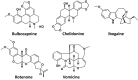
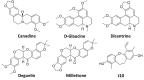
DNA G-quadruplexes (G4s) are key structures for the development of targeted anticancer therapies. In this context, ligands selectively interacting with G4s can represent valuable anticancer drugs. Aiming at speeding up the identification of G4-targeting synthetic or natural compounds, we developed an affinity chromatography-based assay, named G-quadruplex on Oligo Affinity Support (G4-OAS), by synthesizing G4-forming sequences on commercially available polystyrene OAS. Then, due to unspecific binding of several hydrophobic ligands on nude OAS, we moved to Controlled Pore Glass (CPG). We thus conceived an ad hoc functionalized, universal support on which both the on-support elongation and deprotection of the G4-forming oligonucleotides can be performed, along with the successive affinity chromatography-based assay, renamed as G-quadruplex on Controlled Pore Glass (G4-CPG) assay. Here we describe these assays and their applications to the screening of several libraries of chemically different putative G4 ligands. Finally, ongoing studies and outlook of our G4-CPG assay are reported.
Keywords: G-quadruplexes; affinity chromatography; cancer; drug discovery; oligonucleotides.
© 2022 The Authors. Published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
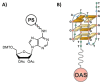


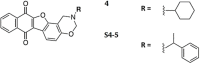

Similar articles
-
Tandem application of ligand-based virtual screening and G4-OAS assay to identify novel G-quadruplex-targeting chemotypes.Biochim Biophys Acta Gen Subj. 2017 May;1861(5 Pt B):1341-1352. doi: 10.1016/j.bbagen.2017.01.024. Epub 2017 Jan 24. Biochim Biophys Acta Gen Subj. 2017. PMID: 28130159
-
G-quadruplex on oligo affinity support (G4-OAS): an easy affinity chromatography-based assay for the screening of G-quadruplex ligands.Anal Chem. 2014 May 6;86(9):4126-30. doi: 10.1021/ac500444m. Epub 2014 Apr 14. Anal Chem. 2014. PMID: 24725064
-
Tailoring a lead-like compound targeting multiple G-quadruplex structures.Eur J Med Chem. 2019 Feb 1;163:295-306. doi: 10.1016/j.ejmech.2018.11.058. Epub 2018 Nov 27. Eur J Med Chem. 2019. PMID: 30529547
-
Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy.Molecules. 2019 Jan 24;24(3):429. doi: 10.3390/molecules24030429. Molecules. 2019. PMID: 30682877 Free PMC article. Review.
-
Targeting G-Quadruplex DNA for Cancer Chemotherapy.Curr Drug Discov Technol. 2022;19(3):e140222201110. doi: 10.2174/1570163819666220214115408. Curr Drug Discov Technol. 2022. PMID: 35156574 Review.
Cited by
-
Exploring the Binding of Natural Compounds to Cancer-Related G-Quadruplex Structures: From 9,10-Dihydrophenanthrenes to Their Dimeric and Glucoside Derivatives.Int J Mol Sci. 2023 Apr 24;24(9):7765. doi: 10.3390/ijms24097765. Int J Mol Sci. 2023. PMID: 37175474 Free PMC article.
-
Selective Targeting of Cancer-Related G-Quadruplex Structures by the Natural Compound Dicentrine.Int J Mol Sci. 2023 Feb 17;24(4):4070. doi: 10.3390/ijms24044070. Int J Mol Sci. 2023. PMID: 36835480 Free PMC article.
-
Insights into the Small Molecule Targeting of Biologically Relevant G-Quadruplexes: An Overview of NMR and Crystal Structures.Pharmaceutics. 2022 Nov 1;14(11):2361. doi: 10.3390/pharmaceutics14112361. Pharmaceutics. 2022. PMID: 36365179 Free PMC article. Review.
-
Combining Cyclic Triimidazo Triazine Core With Ethynyl-N-Methyl-Pyridinium Groups for Targeting G-Quadruplex Structures.Arch Pharm (Weinheim). 2025 Jul;358(7):e70037. doi: 10.1002/ardp.70037. Arch Pharm (Weinheim). 2025. PMID: 40605281 Free PMC article.
-
Optimization of G-Quadruplex Ligands through a SAR Study Combining Parallel Synthesis and Screening of Cationic Bis(acylhydrazones).Chemistry. 2023 Jan 18;29(4):e202202427. doi: 10.1002/chem.202202427. Epub 2022 Nov 30. Chemistry. 2023. PMID: 36286608 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

