Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness
- PMID: 35608456
- PMCID: PMC9161372
- DOI: 10.1126/science.abm1208
Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness
Abstract
Repeated emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants with increased fitness underscores the value of rapid detection and characterization of new lineages. We have developed PyR0, a hierarchical Bayesian multinomial logistic regression model that infers relative prevalence of all viral lineages across geographic regions, detects lineages increasing in prevalence, and identifies mutations relevant to fitness. Applying PyR0 to all publicly available SARS-CoV-2 genomes, we identify numerous substitutions that increase fitness, including previously identified spike mutations and many nonspike mutations within the nucleocapsid and nonstructural proteins. PyR0 forecasts growth of new lineages from their mutational profile, ranks the fitness of lineages as new sequences become available, and prioritizes mutations of biological and public health concern for functional characterization.
Figures
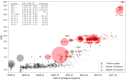

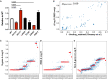
Update of
-
Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness.medRxiv [Preprint]. 2022 Feb 16:2021.09.07.21263228. doi: 10.1101/2021.09.07.21263228. medRxiv. 2022. Update in: Science. 2022 Jun 17;376(6599):1327-1332. doi: 10.1126/science.abm1208. PMID: 35194619 Free PMC article. Updated. Preprint.
References
-
- Davies N. G., Abbott S., Barnard R. C., Jarvis C. I., Kucharski A. J., Munday J. D., Pearson C. A. B., Russell T. W., Tully D. C., Washburne A. D., Wenseleers T., Gimma A., Waites W., Wong K. L. M., van Zandvoort K., Silverman J. D., Diaz-Ordaz K., Keogh R., Eggo R. M., Funk S., Jit M., Atkins K. E., Edmunds W. J.; CMMID COVID-19 Working Group; COVID-19 Genomics UK (COG-UK) Consortium , Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372, eabg3055 (2021). 10.1126/science.abg3055 - DOI - PMC - PubMed
-
- Volz E., Mishra S., Chand M., Barrett J. C., Johnson R., Geidelberg L., Hinsley W. R., Laydon D. J., Dabrera G., O’Toole Á., Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C. V., Boyd O., Loman N. J., McCrone J. T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D. K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D. P., Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N. M.; COVID-19 Genomics UK (COG-UK) consortium , Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593, 266–269 (2021). - PubMed
-
- Stefanelli P., Trentini F., Guzzetta G., Marziano V., Mammone A., Poletti P., Grané C. M., Manica M., del Manso M., Andrianou X., Others, Co-circulation of SARS-CoV-2 variants B. 1.1. 7 and P. 1. medRxiv (2021) (available at https://www.medrxiv.org/content/10.1101/2021.04.06.21254923v1.abstract). - DOI
-
- Stefanelli P., Trentini F., Guzzetta G., Marziano V., Mammone A., Sane Schepisi M., Poletti P., Molina Grané C., Manica M., Del Manso M., Andrianou X., Ajelli M., Rezza G., Brusaferro S., Merler S.; COVID-19 National Microbiology Surveillance Study Group , Co-circulation of SARS-CoV-2 Alpha and Gamma variants in Italy, February and March 2021. Euro Surveill. 27, (2022). 10.2807/1560-7917.ES.2022.27.5.2100429 - DOI - PMC - PubMed
-
- Vöhringer H. S., Sanderson T., Sinnott M., De Maio N., Nguyen T., Goater R., Schwach F., Harrison I., Hellewell J., Ariani C. V., Gonçalves S., Jackson D. K., Johnston I., Jung A. W., Saint C., Sillitoe J., Suciu M., Goldman N., Panovska-Griffiths J., Birney E., Volz E., Funk S., Kwiatkowski D., Chand M., Martincorena I., Barrett J. C., Gerstung M.; Wellcome Sanger Institute COVID-19 Surveillance Team; COVID-19 Genomics UK (COG-UK) Consortium* , Genomic reconstruction of the SARS-CoV-2 epidemic in England. Nature 600, 506–511 (2021). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

