Retrospective Genomic Characterization of a 2017 Dengue Virus Outbreak, Burkina Faso
- PMID: 35608626
- PMCID: PMC9155902
- DOI: 10.3201/eid2806.212491
Retrospective Genomic Characterization of a 2017 Dengue Virus Outbreak, Burkina Faso
Abstract
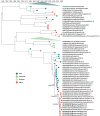
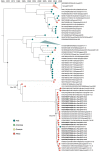
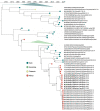
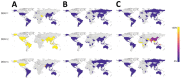
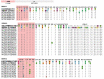
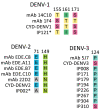
Knowledge of contemporary genetic composition of dengue virus (DENV) in Africa is lacking. By using next-generation sequencing of samples from the 2017 DENV outbreak in Burkina Faso, we isolated 29 DENV genomes (5 serotype 1, 16 serotype 2 [DENV-2], and 8 serotype 3). Phylogenetic analysis demonstrated the endemic nature of DENV-2 in Burkina Faso. We noted discordant diagnostic results, probably related to genetic divergence between these genomes and the Trioplex PCR. Forward and reverse1 primers had a single mismatch when mapped to the DENV-2 genomes, probably explaining the insensitivity of the molecular test. Although we observed considerable homogeneity between the Dengvaxia and TetraVax-DV-TV003 vaccine strains as well as B cell epitopes compared with these genomes, we noted unique divergence. Continual surveillance of dengue virus in Africa is needed to clarify the ongoing novel evolutionary dynamics of circulating virus populations and support the development of effective diagnostic, therapeutic, and preventive countermeasures.
Keywords: Africa; Burkina Faso; RT-PCR; dengue virus; diagnostics; mosquito-borne diseases; next-generation sequencing; outbreak; phylogenetics; vector-borne infections; viruses.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

