Geographic Origin and Vertical Transmission of Leishmania infantum Parasites in Hunting Hounds, United States
- PMID: 35608628
- PMCID: PMC9155895
- DOI: 10.3201/eid2806.211746
Geographic Origin and Vertical Transmission of Leishmania infantum Parasites in Hunting Hounds, United States
Abstract
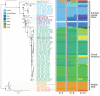
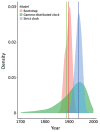
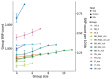
Vertical transmission of leishmaniasis is common but is difficult to study against the background of pervasive vector transmission. We present genomic data from dogs in the United States infected with Leishmania infantum parasites; these infections have persisted in the apparent absence of vector transmission. We demonstrate that these parasites were introduced from the Old World separately and more recently than L. infantum from South America. The parasite population shows unusual genetics consistent with a lack of meiosis: a high level of heterozygous sites shared across all isolates and no decrease in linkage with genomic distance between variants. Our data confirm that this parasite population has been evolving with little or no sexual reproduction. This demonstration of vertical transmission has profound implications for the population genetics of Leishmania parasites. When investigating transmission in complex natural settings, considering vertical transmission alongside vector transmission is vital.
Keywords: Leishmania infantum; United States; clonal evolution; genomics; hunting hounds; leishmaniasis; parasites; vertical transmission.
Figures




References
-
- Bsrat A, Berhe M, Gadissa E, Taddele H, Tekle Y, Hagos Y, et al. Serological investigation of visceral Leishmania infection in human and its associated risk factors in Welkait District, Western Tigray, Ethiopia. Parasite Epidemiol Control. 2018;3:13–20. 10.1016/j.parepi.2017.10.004 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

