Systematic analysis of Kaposi's sarcoma (KS)-associated herpesvirus genomes from a KS case-control study in Cameroon: Evidence of dual infections but no association between viral sequence variation and KS risk
- PMID: 35608873
- PMCID: PMC10043945
- DOI: 10.1002/ijc.34136
Systematic analysis of Kaposi's sarcoma (KS)-associated herpesvirus genomes from a KS case-control study in Cameroon: Evidence of dual infections but no association between viral sequence variation and KS risk
Abstract
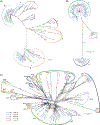
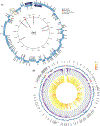
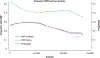
In sub-Saharan Africa, Kaposi's sarcoma-associated herpesvirus (KSHV) is endemic, and Kaposi's sarcoma (KS) is a significant public health problem. Until recently, KSHV genotype analysis was performed using variable gene regions, representing a small fraction of the genome, and thus the contribution of sequence variation to viral transmission or pathogenesis are understudied. We performed near full-length KSHV genome sequence analysis on samples from 43 individuals selected from a large Cameroonian KS case-control study. KSHV genomes were obtained from 21 KS patients and 22 control participants. Phylogenetic analysis of the K1 region indicated the majority of sequences were A5 or B1 subtypes and all three K15 alleles were represented. Unique polymorphisms in the KSHV genome were observed including large gene deletions. We found evidence of multiple distinct KSHV genotypes in three individuals. Additionally, our analyses indicate that recombination is prevalent suggesting that multiple KSHV infections may not be uncommon overall. Most importantly, a detailed analysis of KSHV genomes from KS patients and control participants did not find a correlation between viral sequence variations and disease. Our study is the first to systematically compare near full-length KSHV genome sequences between KS cases and controls in the same endemic region to identify possible sequence variations associated with disease risk.
Keywords: Cameroon; Kaposi's sarcoma; Kaposi's sarcoma-associated herpesvirus; human herpesvirus 8; next-generation sequencing; whole genome sequencing.
© 2022 UICC.
Conflict of interest statement
CONFLICT OF INTEREST
Dr. Neneh Salah is currently an employee of GSK and owns company shares. No other authors have COI to declare.
Figures


Similar articles
-
Whole-Genome Sequencing of Kaposi's Sarcoma-Associated Herpesvirus from Zambian Kaposi's Sarcoma Biopsy Specimens Reveals Unique Viral Diversity.J Virol. 2015 Dec;89(24):12299-308. doi: 10.1128/JVI.01712-15. Epub 2015 Sep 30. J Virol. 2015. PMID: 26423952 Free PMC article. Clinical Trial.
-
Diverse genotypes of Kaposi's sarcoma associated herpesvirus (KSHV) identified in infant blood infections in African childhood-KS and HIV/AIDS endemic region.J Med Virol. 2007 Oct;79(10):1555-61. doi: 10.1002/jmv.20952. J Med Virol. 2007. PMID: 17705172 Free PMC article.
-
Genotypic analysis at multiple loci across Kaposi's sarcoma herpesvirus (KSHV) DNA molecules: clustering patterns, novel variants and chimerism.J Clin Virol. 2002 Jan;23(3):119-48. doi: 10.1016/s1386-6532(01)00205-0. J Clin Virol. 2002. PMID: 11595592
-
Kaposi's Sarcoma-Associated Herpesvirus: Epidemiology and Molecular Biology.Adv Exp Med Biol. 2017;1018:91-127. doi: 10.1007/978-981-10-5765-6_7. Adv Exp Med Biol. 2017. PMID: 29052134 Review.
-
Kaposi's Sarcoma (KS)-associated herpesvirus and its role in KS.Infect Agents Dis. 1996 Oct;5(4):215-22. Infect Agents Dis. 1996. PMID: 8884366 Review.
Cited by
-
HIV-associated cancers and lymphoproliferative disorders caused by Kaposi sarcoma herpesvirus and Epstein-Barr virus.Clin Microbiol Rev. 2024 Sep 12;37(3):e0002223. doi: 10.1128/cmr.00022-23. Epub 2024 Jun 20. Clin Microbiol Rev. 2024. PMID: 38899877 Free PMC article. Review.
-
Genome evolution of Kaposi sarcoma-associated herpesvirus (KSHV).J Virol. 2025 May 20;99(5):e0195024. doi: 10.1128/jvi.01950-24. Epub 2025 Apr 16. J Virol. 2025. PMID: 40237497 Free PMC article.
-
Sequencing of Kaposi's Sarcoma Herpesvirus (KSHV) genomes from persons of diverse ethnicities and provenances with KSHV-associated diseases demonstrate multiple infections, novel polymorphisms, and low intra-host variance.PLoS Pathog. 2024 Jul 15;20(7):e1012338. doi: 10.1371/journal.ppat.1012338. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39008527 Free PMC article.
-
Evolutionary analysis and population dynamics in the global transmission of Kaposi's sarcoma-associated herpesvirus.Arch Virol. 2025 Mar 27;170(5):92. doi: 10.1007/s00705-025-06259-9. Arch Virol. 2025. PMID: 40146465 Free PMC article.
-
Viral Load Measurements for Kaposi Sarcoma Herpesvirus (KSHV/HHV8): Review and an Updated Assay.J Med Virol. 2024 Dec;96(12):e70105. doi: 10.1002/jmv.70105. J Med Virol. 2024. PMID: 39648698 Review.
References
-
- Parkin DM, Hammerl L, Ferlay J, Kantelhardt EJ. Cancer in Africa 2018: the role of infections. Int J Cancer. 2020;146:2089–2103. - PubMed
-
- Sankaranarayanan R, Swaminathan R, World Health Organization, International Agency for Research on Cancer. Cancer Survival in Africa, Asia, the Caribbean and Central America. Lyon, France: International Agency for Research on Cancer, World Health Organization; 2011.
-
- Chen WC, Singh E, Muchengeti M, et al. Johannesburg cancer study (JCS): contribution to knowledge and opportunities arising from 20 years of data collection in an African setting. Cancer Epidemiol. 2020; 65:101701. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

