Combining datasets for maize root seedling traits increases the power of GWAS and genomic prediction accuracies
- PMID: 35608947
- PMCID: PMC9467658
- DOI: 10.1093/jxb/erac236
Combining datasets for maize root seedling traits increases the power of GWAS and genomic prediction accuracies
Abstract
The identification of genomic regions associated with root traits and the genomic prediction of untested genotypes can increase the rate of genetic gain in maize breeding programs targeting roots traits. Here, we combined two maize association panels with different genetic backgrounds to identify single nucleotide polymorphisms (SNPs) associated with root traits, and used a genome-wide association study (GWAS) and to assess the potential of genomic prediction for these traits in maize. For this, we evaluated 377 lines from the Ames panel and 302 from the Backcrossed Germplasm Enhancement of Maize (BGEM) panel in a combined panel of 679 lines. The lines were genotyped with 232 460 SNPs, and four root traits were collected from 14-day-old seedlings. We identified 30 SNPs significantly associated with root traits in the combined panel, whereas only two and six SNPs were detected in the Ames and BGEM panels, respectively. Those 38 SNPs were in linkage disequilibrium with 35 candidate genes. In addition, we found higher prediction accuracy in the combined panel than in the Ames or BGEM panel. We conclude that combining association panels appears to be a useful strategy to identify candidate genes associated with root traits in maize and improve the efficiency of genomic prediction.
Keywords: Zea mays L; Association mapping; candidate genes; genomic selection; inbred line panel; linkage disequilibrium; population structure.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures


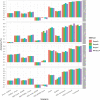
References
-
- Abdel-Ghani AH, Kumar B, Reyes-Matamoros J, Gonzalez-Portilla PJ, Jansen C, San Martin JP, Lee M, Lubberstedt T.. 2013. Genotypic variation and relationships between seedling and adult plant traits in maize (Zea mays L.) inbred lines grown under contrasting nitrogen levels. Euphytica 189, 123–133. - PubMed
-
- Andorf C, Beavis WD, Hufford M, Smith S, Suza W, Wang K, Woodhouse M, Yu J, Lübberstedt T.. 2019. Technological advances in maize breeding: past, present and future. Theoretical and Applied Genetics 132, 817–849. - PubMed
-
- Bates D, Maechler M, Bolker B, Walker S.. 2015. Fitting linear mixed-effects models using ‘lme4’. Journal of Statistical Software 67, 1–48.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

