Biogenesis and Regulatory Roles of Circular RNAs
- PMID: 35609906
- PMCID: PMC10119891
- DOI: 10.1146/annurev-cellbio-120420-125117
Biogenesis and Regulatory Roles of Circular RNAs
Abstract
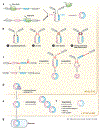
Covalently closed, single-stranded circular RNAs can be produced from viral RNA genomes as well as from the processing of cellular housekeeping noncoding RNAs and precursor messenger RNAs. Recent transcriptomic studies have surprisingly uncovered that many protein-coding genes can be subjected to backsplicing, leading to widespread expression of a specific type of circular RNAs (circRNAs) in eukaryotic cells. Here, we discuss experimental strategies used to discover and characterize diverse circRNAs at both the genome and individual gene scales. We further highlight the current understanding of how circRNAs are generated and how the mature transcripts function. Some circRNAs act as noncoding RNAs to impact gene regulation by serving as decoys or competitors for microRNAs and proteins. Others form extensive networks of ribonucleoprotein complexes or encode functional peptides that are translated in response to certain cellular stresses. Overall, circRNAs have emerged as an important class of RNAmolecules in gene expression regulation that impact many physiological processes, including early development, immune responses, neurogenesis, and tumorigenesis.
Keywords: backsplicing; circRNA; microRNA; noncoding RNA; pre-mRNA splicing; translation.
Figures
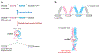
References
-
- Aktaş T, Avşar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, et al. 2017. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544:115–19 - PubMed
-
- Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. 1980. Some yeast mitochondrial RNAs are circular. Cell 19:313–19 - PubMed
-
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, et al. 2014. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56:55–66 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

