Extrachromosomal DNA in Cancer
- PMID: 35609926
- PMCID: PMC10508221
- DOI: 10.1146/annurev-genom-120821-100535
Extrachromosomal DNA in Cancer
Abstract
In cancer, complex genome rearrangements and other structural alterations, including the amplification of oncogenes on circular extrachromosomal DNA (ecDNA) elements, drive the formation and progression of tumors. ecDNA is a particularly challenging structural alteration. By untethering oncogenes from chromosomal constraints, it elevates oncogene copy number, drives intratumoral genetic heterogeneity, promotes rapid tumor evolution, and results in treatment resistance. The profound changes in DNA shape and nuclear architecture generated by ecDNA alter the transcriptional landscape of tumors by catalyzing new types of regulatory interactions that do not occur on chromosomes. The current suite of tools for interrogating cancer genomes is well suited for deciphering sequence but has limited ability to resolve the complex changes in DNA structure and dynamics that ecDNA generates. Here, we review the challenges of resolving ecDNA form and function and discuss the emerging tool kit for deciphering ecDNA architecture and spatial organization, including what has been learned to date about how this dramatic change in shape alters tumor development, progression, and drug resistance.
Keywords: HSRs; cancer; ecDNA; ecDNA evolution; ecDNA hubs; enhancer hijacking; homogeneously staining regions.
Conflict of interest statement
DISCLOSURE STATEMENT
VB is a co-founder, consultant, SAB member and has equity interest in Boundless Bio, inc. and Abterra, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. P.S.M. is a co-founder of Boundless Bio, Inc. He chairs the Scientific Advisory Board, for which he is compensated.
Figures


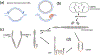
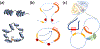
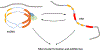
References
-
- Adelman K, Martin BJE. 2021. ecDNA party bus: Bringing the enhancer to you. Mol Cell 81(9):1866–1867 - PubMed
-
- Alt FW, Kellems RE, Bertino JR, Schimke RT. 1978. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. Journal of Biological Chemistry 253(5):1357–1370 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

