Mast cell marker gene signature in head and neck squamous cell carcinoma
- PMID: 35610596
- PMCID: PMC9128261
- DOI: 10.1186/s12885-022-09673-3
Mast cell marker gene signature in head and neck squamous cell carcinoma
Abstract
Background: Mast cells can reshape the tumour immune microenvironment and greatly affect tumour occurrence and development. However, mast cell gene prognostic and predictive value in head and neck squamous cell carcinoma (HNSCC) remains unclear. This study was conducted to identify and establish a prognostic mast cell gene signature (MCS) for assessing the prognosis and immunotherapy response of patients with HNSCC.
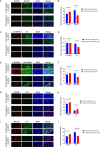
Methods: Mast cell marker genes in HNSCC were identified using single-cell RNA sequencing analysis. A dataset from The Cancer Genome Atlas was divided into a training cohort to construct the MCS model and a testing cohort to validate the model. Fluorescence in-situ hybridisation was used to evaluate the MCS model gene expression in tissue sections from patients with HNSCC who had been treated with programmed cell death-1 inhibitors and further validate the MCS.
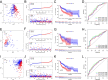
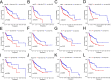
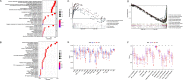
Results: A prognostic MCS comprising nine genes (KIT, RAB32, CATSPER1, SMYD3, LINC00996, SOCS1, AP2M1, LAT, and HSP90B1) was generated by comprehensively analysing clinical features and 47 mast cell-related genes. The MCS effectively distinguished survival outcomes across the training, testing, and entire cohorts as an independent prognostic factor. Furthermore, we identified patients with favourable immune cell infiltration status and immunotherapy responses. Fluorescence in-situ hybridisation supported the MCS immunotherapy response of patients with HNSCC prediction, showing increased high-risk gene expression and reduced low-risk gene expression in immunotherapy-insensitive patients.
Conclusions: Our MCS provides insight into the roles of mast cells in HNSCC prognosis and may have applications as an immunotherapy response predictive indicator in patients with HNSCC and a reference for immunotherapy decision-making.
Keywords: Head and neck squamous cell carcinoma; Immune infiltration; Immunotherapy; Risk score; Single-cell RNA sequencing; Tumour microenvironment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

