Identification of circ_0000375 and circ_0011536 as novel diagnostic biomarkers of colorectal cancer
- PMID: 35611198
- PMCID: PMC9048569
- DOI: 10.12998/wjcc.v10.i11.3352
Identification of circ_0000375 and circ_0011536 as novel diagnostic biomarkers of colorectal cancer
Abstract
Background: Colorectal cancer (CRC) imposes a tremendous burden on human health, with high morbidity and mortality. Circular ribonucleic acids (circRNAs), a new type of noncoding RNA, are considered to participate in cancer pathogenesis as microRNA (miRNA) sponges. However, the dysregulation and biological functions of circRNAs in CRC remain to be explored.
Aim: To identify potential circRNA biomarkers of CRC and explore their functions in CRC carcinogenesis.
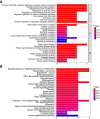
Methods: CircRNAs and miRNAs differentially expressed in CRC tissues were identified by analyzing expression profiles from the Gene Expression Omnibus (GEO) database. Circ_0000375 and circ_0011536 were selected as CRC biomarker candidates. Quantitative real-time polymerase chain reaction was utilized to evaluate the expression of these 2 circRNAs in CRC tissues, serums and cell lines. Receiver operating characteristic curves were generated to assess the diagnostic performances of these 2 circRNAs. Then, functional experiments, including cell counting kit-8, wound healing and Transwell invasion assays, were performed after the overexpression of circ_0000375 and circ_0011536 in CRC cell lines. Furthermore, candidate target miRNAs of circ_0000375 and circ_0011536 were predicted via bioinformatics analysis. The expression levels of these miRNAs were explored in CRC cell lines and tissues from GEO datasets. A luciferase reporter assay was developed to examine the interactions between circRNAs and miRNAs. Based on the target miRNAs and downstream genes, functional enrichment analyses were applied to reveal the critical signaling pathways involved in CRC carcinogenesis.
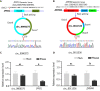
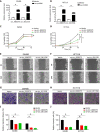
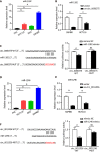
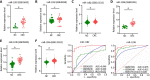
Results: Downregulated circ_0000375 and circ_0011536 expression was observed in CRC tissues in GSE126095, clinical CRC tissue and serum samples and CRC cell lines. The areas under the curve for circ_0000375 and circ_0011536 were 0.911 and 0.885 in CRC tissue and 0.976 and 0.982 in CRC serum, respectively. Moreover, the serum levels of these 2 circRNAs were higher in patients at 30 d postsurgery than in patients before surgery, suggesting that the serum expression of circ_0000375 and circ_0011536 is related to CRC tumorigenesis. Circ_0000375 and circ_0011536 overexpression inhibited the proliferation, migration and invasion of CRC cells. Furthermore, miR-1182 and miR-1246, which were overexpressed in CRC tissues in GSE41655, GSE49246 and GSE115513, were verified as target miRNAs of circ_0000375 and circ_0011536, respectively, by luciferase reporter assays. The downstream genes of miR-1182 and miR-1246 were enriched in some CRC-associated pathways, such as the Wnt signaling pathway.
Conclusion: Circ_0000375 and circ_0011536 may function as tumor suppressors in CRC progression, serving as novel biomarkers for CRC diagnosis and as promising candidates for therapeutic exploration.
Keywords: Biomarker; Cir_0000375; Circ_0011536; Colorectal cancer; MicroRNA; Tumor suppressor.
©The Author(s) 2022. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: There are no conflicts of interest to report.
Figures


References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin . 2021;71:209–249. - PubMed
-
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin . 2020;70:145–164. - PubMed
-
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet . 2014;383:1490–1502. - PubMed
LinkOut - more resources
Full Text Sources

