miR-223 Exerts Translational Control of Proatherogenic Genes in Macrophages
- PMID: 35611698
- PMCID: PMC9213086
- DOI: 10.1161/CIRCRESAHA.121.319120
miR-223 Exerts Translational Control of Proatherogenic Genes in Macrophages
Abstract
Background: A significant burden of atherosclerotic disease is driven by inflammation. Recently, microRNAs (miRNAs) have emerged as important factors driving and protecting from atherosclerosis. miR-223 regulates cholesterol metabolism and inflammation via targeting both cholesterol biosynthesis pathway and NFkB signaling pathways; however, its role in atherosclerosis has not been investigated. We hypothesize that miR-223 globally regulates core inflammatory pathways in macrophages in response to inflammatory and atherogenic stimuli thus limiting the progression of atherosclerosis.
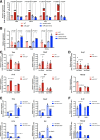
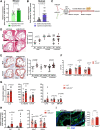
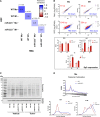
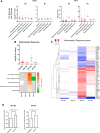
Methods and results: Loss of miR-223 in macrophages decreases Abca1 gene and protein expression as well as cholesterol efflux to apoA1 (Apolipoprotein A1) and enhances proinflammatory gene expression. In contrast, overexpression of miR-223 promotes the efflux of cholesterol and macrophage polarization toward an anti-inflammatory phenotype. These beneficial effects of miR-223 are dependent on its target gene, the transcription factor Sp3. Consistent with the antiatherogenic effects of miR-223 in vitro, mice receiving miR223-/- bone marrow exhibit increased plaque size, lipid content, and circulating inflammatory cytokines (ie, IL-1β). Deficiency of miR-223 in bone marrow-derived cells also results in an increase in circulating pro-atherogenic cells (total monocytes and neutrophils) compared with control mice. Furthermore, the expression of miR-223 target gene (Sp3) and pro-inflammatory marker (Il-6) are enhanced whereas the expression of Abca1 and anti-inflammatory marker (Retnla) are reduced in aortic arches from mice lacking miR-223 in bone marrow-derived cells. In mice fed a high-cholesterol diet and in humans with unstable carotid atherosclerosis, the expression of miR-223 is increased. To further understand the molecular mechanisms underlying the effect of miR-223 on atherosclerosis in vivo, we characterized global RNA translation profile of macrophages isolated from mice receiving wild-type or miR223-/- bone marrow. Using ribosome profiling, we reveal a notable upregulation of inflammatory signaling and lipid metabolism at the translation level but less significant at the transcription level. Analysis of upregulated genes at the translation level reveal an enrichment of miR-223-binding sites, confirming that miR-223 exerts significant changes in target genes in atherogenic macrophages via altering their translation.
Conclusions: Our study demonstrates that miR-223 can protect against atherosclerosis by acting as a global regulator of RNA translation of cholesterol efflux and inflammation pathways.
Keywords: animal models of human disease; cholesterol; inflammation; lipids; metabolism; vascular biology.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

