ALTEN: A High-Fidelity Primary Tissue-Engineering Platform to Assess Cellular Responses Ex Vivo
- PMID: 35611998
- PMCID: PMC9313544
- DOI: 10.1002/advs.202103332
ALTEN: A High-Fidelity Primary Tissue-Engineering Platform to Assess Cellular Responses Ex Vivo
Abstract
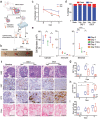
To fully investigate cellular responses to stimuli and perturbations within tissues, it is essential to replicate the complex molecular interactions within the local microenvironment of cellular niches. Here, the authors introduce Alginate-based tissue engineering (ALTEN), a biomimetic tissue platform that allows ex vivo analysis of explanted tissue biopsies. This method preserves the original characteristics of the source tissue's cellular milieu, allowing multiple and diverse cell types to be maintained over an extended period of time. As a result, ALTEN enables rapid and faithful characterization of perturbations across specific cell types within a tissue. Importantly, using single-cell genomics, this approach provides integrated cellular responses at the resolution of individual cells. ALTEN is a powerful tool for the analysis of cellular responses upon exposure to cytotoxic agents and immunomodulators. Additionally, ALTEN's scalability using automated microfluidic devices for tissue encapsulation and subsequent transport, to enable centralized high-throughput analysis of samples gathered by large-scale multicenter studies, is shown.
Keywords: alginate; ex vivo drug screening; single-cell RNAseq; three dimensional culture; tissue microenvironment; whole-tissue organoids.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Hay M., Thomas D. W., Craighead J. L., Economides C., Rosenthal J., Nat. Biotechnol. 2014, 32, 40. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
