Identification and characterization of key residues in Zika virus envelope protein for virus assembly and entry
- PMID: 35612559
- PMCID: PMC9196690
- DOI: 10.1080/22221751.2022.2082888
Identification and characterization of key residues in Zika virus envelope protein for virus assembly and entry
Abstract
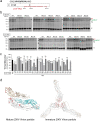
Zika virus (ZIKV), a family member in the Flavivirus genus, has re-emerged as a global public health concern. The envelope (E) proteins of flaviviruses play a dual role in viral assembly and entry. To identify the key residues of E in virus entry, we generated a ZIKV trans-complemented particle (ZIKVTCP) system, in which a subgenomic reporter replicon was packaged by trans-complementation with expression of CprME. We performed mutagenesis studies of the loop regions that protrude from the surface of the virion in the E ectodomains (DI, DII, DIII). Most mutated ZIKVTCPs exhibited deficient egress. Mutations in DII and in the hinge region of DI and DIII affected prM expression. With a bioorthogonal system, photocrosslinking experiments identified crosslinked intracellular E trimers and demonstrated that egress-deficient mutants in DIII impaired E trimerization. Of these mutants, an E-trimerization-dead mutation D389A that nears the E-E interface between two neighbouring spikes in the immature virion completely abolished viral egress. Several mutations abolished ZIKVTCPs' entry, without severely affecting viral egress. Further virus binding experiments demonstrated a deficiency of the mutated ZIKVTCPs in virus attachment. Strikingly, synthesized peptide containing residues of two mutants (268-273aa in DII) could bind to host cells and significantly compete for viral attachment and interfere with viral infection, suggesting an important role of these resides in virus entry. Our findings uncovered the requirement for DIII mediated-E trimerization in viral egress, and discovered a key residue group in DII that participates in virus entry.
Keywords: Zika virus (ZIKV); envelope protein; flavivirus; glycoprotein; mutagenesis; transcomplemented particle system; viral egress; viral entry.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





Similar articles
-
Zika Virus NS2A-Mediated Virion Assembly.mBio. 2019 Oct 29;10(5):e02375-19. doi: 10.1128/mBio.02375-19. mBio. 2019. PMID: 31662457 Free PMC article.
-
Structurally Conserved Domains between Flavivirus and Alphavirus Fusion Glycoproteins Contribute to Replication and Infectious-Virion Production.J Virol. 2022 Jan 26;96(2):e0177421. doi: 10.1128/JVI.01774-21. Epub 2021 Nov 10. J Virol. 2022. PMID: 34757841 Free PMC article.
-
Zika virus structural biology and progress in vaccine development.Biotechnol Adv. 2018 Jan-Feb;36(1):47-53. doi: 10.1016/j.biotechadv.2017.09.004. Epub 2017 Sep 12. Biotechnol Adv. 2018. PMID: 28916391 Review.
-
The Envelope Residues E152/156/158 of Zika Virus Influence the Early Stages of Virus Infection in Human Cells.Cells. 2019 Nov 15;8(11):1444. doi: 10.3390/cells8111444. Cells. 2019. PMID: 31731738 Free PMC article.
-
Structures of Zika Virus E & NS1: Relations with Virus Infection and Host Immune Responses.Adv Exp Med Biol. 2018;1062:77-87. doi: 10.1007/978-981-10-8727-1_6. Adv Exp Med Biol. 2018. PMID: 29845526 Review.
Cited by
-
Broad-spectrum activity against mosquito-borne flaviviruses achieved by a targeted protein degradation mechanism.Nat Commun. 2024 Jun 19;15(1):5179. doi: 10.1038/s41467-024-49161-9. Nat Commun. 2024. PMID: 38898037 Free PMC article.
-
The Evolving Role of Zika Virus Envelope Protein in Viral Entry and Pathogenesis.Viruses. 2025 Jun 6;17(6):817. doi: 10.3390/v17060817. Viruses. 2025. PMID: 40573410 Free PMC article. Review.
-
The Dual Role of TRIM7 in Viral Infections.Viruses. 2024 Aug 12;16(8):1285. doi: 10.3390/v16081285. Viruses. 2024. PMID: 39205259 Free PMC article. Review.
References
-
- Gould EA, Solomon T.. Pathogenic flaviviruses. Lancet. 2008 Feb 9;371(9611):500–509. - PubMed
-
- Dick GW, Kitchen SF, Haddow AJ, et al. . Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952 Sep;46(5):509–520. - PubMed
-
- Musso D, Ko AI, Baud D.. Zika virus infection – after the pandemic. N Engl J Med. 2019 Oct 10;381(15):1444–1457. - PubMed
-
- Lindenbach BD, Thiel H-J, Rice CM.. Flaviviridae: the viruses and their relication. In Knipe DM, Howley PM, (eds.), Fields virology, 5th ed. Philadelphia: Lippincott, Williams & Wilkins; 2007Jan 01:1101–1152.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
