Probing function in ligand-gated ion channels without measuring ion transport
- PMID: 35612603
- PMCID: PMC9136306
- DOI: 10.1085/jgp.202213082
Probing function in ligand-gated ion channels without measuring ion transport
Abstract
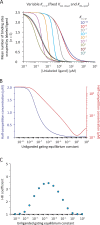
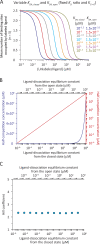
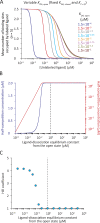
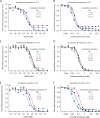
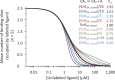
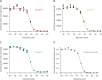
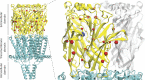
Although the functional properties of ion channels are most accurately assessed using electrophysiological approaches, a number of experimental situations call for alternative methods. Here, working on members of the pentameric ligand-gated ion channel (pLGIC) superfamily, we focused on the practical implementation of, and the interpretation of results from, equilibrium-type ligand-binding assays. Ligand-binding studies of pLGICs are by no means new, but the lack of uniformity in published protocols, large disparities between the results obtained for a given parameter by different groups, and a general disregard for constraints placed on the experimental observations by simple theoretical considerations suggested that a thorough analysis of this classic technique was in order. To this end, we present a detailed practical and theoretical study of this type of assay using radiolabeled α-bungarotoxin, unlabeled small-molecule cholinergic ligands, the human homomeric α7-AChR, and extensive calculations in the framework of a realistic five-binding-site reaction scheme. Furthermore, we show examples of the practical application of this method to tackle two longstanding questions in the field: our results suggest that ligand-binding affinities are insensitive to binding-site occupancy and that mutations to amino-acid residues in the transmembrane domain are unlikely to affect the channel's affinities for ligands that bind to the extracellular domain.
© 2022 Godellas and Grosman.
Figures











Comment in
-
The surprising difficulty of "simple" equilibrium binding measurements on ligand-gated ion channels.J Gen Physiol. 2022 Jul 4;154(7):e202213177. doi: 10.1085/jgp.202213177. Epub 2022 Jun 2. J Gen Physiol. 2022. PMID: 35653137 Free PMC article.
Similar articles
-
Crystallographic studies of pharmacological sites in pentameric ligand-gated ion channels.Biochim Biophys Acta. 2015 Mar;1850(3):511-23. doi: 10.1016/j.bbagen.2014.05.007. Epub 2014 May 14. Biochim Biophys Acta. 2015. PMID: 24836522 Review.
-
Photoaffinity Labeling of Pentameric Ligand-Gated Ion Channels: A Proteomic Approach to Identify Allosteric Modulator Binding Sites.Methods Mol Biol. 2017;1598:157-197. doi: 10.1007/978-1-4939-6952-4_7. Methods Mol Biol. 2017. PMID: 28508361
-
Structural basis for allosteric transitions of a multidomain pentameric ligand-gated ion channel.Proc Natl Acad Sci U S A. 2020 Jun 16;117(24):13437-13446. doi: 10.1073/pnas.1922701117. Epub 2020 Jun 1. Proc Natl Acad Sci U S A. 2020. PMID: 32482881 Free PMC article.
-
Structural basis for the modulation of pentameric ligand-gated ion channel function by lipids.Biochim Biophys Acta Biomembr. 2020 Sep 1;1862(9):183304. doi: 10.1016/j.bbamem.2020.183304. Epub 2020 Apr 18. Biochim Biophys Acta Biomembr. 2020. PMID: 32311340 Review.
-
Common binding sites for cholesterol and neurosteroids on a pentameric ligand-gated ion channel.Biochim Biophys Acta Mol Cell Biol Lipids. 2019 Feb;1864(2):128-136. doi: 10.1016/j.bbalip.2018.11.005. Epub 2018 Nov 22. Biochim Biophys Acta Mol Cell Biol Lipids. 2019. PMID: 30471426 Free PMC article.
Cited by
-
The surprising difficulty of "simple" equilibrium binding measurements on ligand-gated ion channels.J Gen Physiol. 2022 Jul 4;154(7):e202213177. doi: 10.1085/jgp.202213177. Epub 2022 Jun 2. J Gen Physiol. 2022. PMID: 35653137 Free PMC article.
-
An experimental test of the nicotinic hypothesis of COVID-19.Proc Natl Acad Sci U S A. 2022 Nov;119(44):e2204242119. doi: 10.1073/pnas.2204242119. Epub 2022 Oct 24. Proc Natl Acad Sci U S A. 2022. PMID: 36279466 Free PMC article.
-
The Cranefield Awards 2022.J Gen Physiol. 2023 Apr 3;155(4):e202313385. doi: 10.1085/jgp.202313385. Epub 2023 Mar 17. J Gen Physiol. 2023. PMID: 36929935 Free PMC article. No abstract available.
References
-
- Bertrand, S., Devillers-Thiéry A., Palma E., Buisson B., Edelstein S.J., Corringer P.J., Changeux J.P., and Bertrand D.. 1997. Paradoxical allosteric effects of competitive inhibitors on neuronal alpha7 nicotinic receptor mutants. Neuroreport. 8:3591–3596. 10.1097/00001756-199711100-00034 - DOI - PubMed
-
- Blount, P., and Merlie J.P.. 1988. Native folding of an acetylcholine receptor alpha subunit expressed in the absence of other receptor subunits. J. Biol. Chem. 263:1072–1080 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

