Demethylation and Up-Regulation of an Oncogene after Hypomethylating Therapy
- PMID: 35613022
- PMCID: PMC9514878
- DOI: 10.1056/NEJMoa2119771
Demethylation and Up-Regulation of an Oncogene after Hypomethylating Therapy
Abstract
Background: Although hypomethylating agents are currently used to treat patients with cancer, whether they can also reactivate and up-regulate oncogenes is not well elucidated.
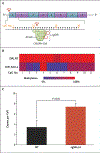
Methods: We examined the effect of hypomethylating agents on SALL4, a known oncogene that plays an important role in myelodysplastic syndrome and other cancers. Paired bone marrow samples that were obtained from two cohorts of patients with myelodysplastic syndrome before and after treatment with a hypomethylating agent were used to explore the relationships among changes in SALL4 expression, treatment response, and clinical outcome. Leukemic cell lines with low or undetectable SALL4 expression were used to study the relationship between SALL4 methylation and expression. A locus-specific demethylation technology, CRISPR-DNMT1-interacting RNA (CRISPR-DiR), was used to identify the CpG island that is critical for SALL4 expression.
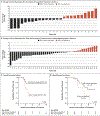
Results: SALL4 up-regulation after treatment with hypomethylating agents was observed in 10 of 25 patients (40%) in cohort 1 and in 13 of 43 patients (30%) in cohort 2 and was associated with a worse outcome. Using CRISPR-DiR, we discovered that demethylation of a CpG island within the 5' untranslated region was critical for SALL4 expression. In cell lines and patients, we confirmed that treatment with a hypomethylating agent led to demethylation of the same CpG region and up-regulation of SALL4 expression.
Conclusions: By combining analysis of patient samples with CRISPR-DiR technology, we found that demethylation and up-regulation of an oncogene after treatment with a hypomethylating agent can indeed occur and should be further studied. (Funded by Associazione Italiana per la Ricerca sul Cancro and others.).
Copyright © 2022 Massachusetts Medical Society.
Conflict of interest statement
Disclosure forms provided by the authors are available with the full text of this article at
Figures

Comment in
-
Oncogene Up-Regulation after Hypomethylating Therapy.N Engl J Med. 2022 Aug 4;387(5):476. doi: 10.1056/NEJMc2208134. N Engl J Med. 2022. PMID: 35921465 No abstract available.
-
Curse and blessing: upregulation of oncogenes upon treatment with hypomethylating agents in myelodysplastic syndrome.Signal Transduct Target Ther. 2022 Aug 9;7(1):276. doi: 10.1038/s41392-022-01143-3. Signal Transduct Target Ther. 2022. PMID: 35945228 Free PMC article. No abstract available.
References
-
- Taylor SM, Jones PA. Mechanism of action of eukaryotic DNA methyltransferase: use of 5-azacytosine-containing DNA. J Mol Biol 1982;162:679–92. - PubMed
-
- Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 1992;71:865–73. - PubMed
-
- Bender CM, Zingg JM, Jones PA. DNA methylation as a target for drug design. Pharm Res 1998;15:175–87. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
