Thalamocortical axons regulate neurogenesis and laminar fates in the early sensory cortex
- PMID: 35613048
- PMCID: PMC9295754
- DOI: 10.1073/pnas.2201355119
Thalamocortical axons regulate neurogenesis and laminar fates in the early sensory cortex
Abstract
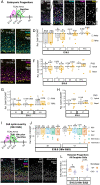
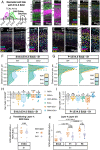
Area-specific axonal projections from the mammalian thalamus shape unique cellular organization in target areas in the adult neocortex. How these axons control neurogenesis and early neuronal fate specification is poorly understood. By using mutant mice lacking the majority of thalamocortical axons, we show that these axons are required for the production and specification of the proper number of layer 4 neurons in primary sensory areas by the neonatal stage. Part of these area-specific roles is played by the thalamus-derived molecule, VGF. Our work reveals that extrinsic cues from sensory thalamic projections have an early role in the formation of cortical cytoarchitecture by enhancing the production and specification of layer 4 neurons.
Keywords: cell fate; neocortex; neurogenesis; thalamocortical.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
Thalamocortical axons control the cytoarchitecture of neocortical layers by area-specific supply of VGF.Elife. 2022 Mar 15;11:e67549. doi: 10.7554/eLife.67549. Elife. 2022. PMID: 35289744 Free PMC article.
-
Development of the thalamus: From early patterning to regulation of cortical functions.Wiley Interdiscip Rev Dev Biol. 2019 Sep;8(5):e345. doi: 10.1002/wdev.345. Epub 2019 Apr 29. Wiley Interdiscip Rev Dev Biol. 2019. PMID: 31034163 Review.
-
Interactions between growing thalamocortical afferent axons and the neocortical primordium in normal and reeler mutant mice.Anat Embryol (Berl). 1994 Aug;190(2):137-54. doi: 10.1007/BF00193411. Anat Embryol (Berl). 1994. PMID: 7818087
-
Laminar specific attachment and neurite outgrowth of thalamic neurons on cultured slices of developing cerebral neocortex.Development. 1994 Oct;120(10):2811-22. doi: 10.1242/dev.120.10.2811. Development. 1994. PMID: 7607072
-
Inputs from the thalamocortical system on axon pathfinding mechanisms.Curr Opin Neurobiol. 2014 Aug;27:143-50. doi: 10.1016/j.conb.2014.03.013. Epub 2014 Apr 17. Curr Opin Neurobiol. 2014. PMID: 24742382 Review.
Cited by
-
Fate plasticity of interneuron specification.iScience. 2025 Mar 27;28(4):112295. doi: 10.1016/j.isci.2025.112295. eCollection 2025 Apr 18. iScience. 2025. PMID: 40264797 Free PMC article.
-
Plasticity of thalamocortical axons is regulated by serotonin levels modulated by preterm birth.Proc Natl Acad Sci U S A. 2023 Aug 15;120(33):e2301644120. doi: 10.1073/pnas.2301644120. Epub 2023 Aug 7. Proc Natl Acad Sci U S A. 2023. PMID: 37549297 Free PMC article.
-
Neurogenesis in primates versus rodents and the value of non-human primate models.Natl Sci Rev. 2023 Sep 15;10(11):nwad248. doi: 10.1093/nsr/nwad248. eCollection 2023 Nov. Natl Sci Rev. 2023. PMID: 38025664 Free PMC article. Review.
-
Synaptic plasticity in human thalamocortical assembloids.bioRxiv [Preprint]. 2024 Mar 12:2024.02.01.578421. doi: 10.1101/2024.02.01.578421. bioRxiv. 2024. Update in: Cell Rep. 2024 Aug 27;43(8):114503. doi: 10.1016/j.celrep.2024.114503. PMID: 38352415 Free PMC article. Updated. Preprint.
-
Evaluation of advances in cortical development using model systems.Dev Neurobiol. 2022 Jul;82(5):408-427. doi: 10.1002/dneu.22879. Epub 2022 May 29. Dev Neurobiol. 2022. PMID: 35644985 Free PMC article. Review.
References
-
- McConnell S. K., The determination of neuronal fate in the cerebral cortex. Trends Neurosci. 12, 342–349 (1989). - PubMed
-
- Oberst P., et al. , Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature 573, 370–374 (2019). - PubMed
-
- Peters A., Jones E. G., Cerebral Cortex: Cellular Components of the Cerebral Cortex (Springer, 1984).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

