An adapted consensus protein design strategy for identifying globally optimal biotherapeutics
- PMID: 35613320
- PMCID: PMC9135432
- DOI: 10.1080/19420862.2022.2073632
An adapted consensus protein design strategy for identifying globally optimal biotherapeutics
Abstract
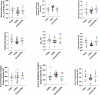
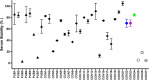
Biotherapeutic optimization, whether to improve general properties or to engineer specific attributes, is a time-consuming process with uncertain outcomes. Conversely, Consensus Protein Design has been shown to be a viable approach to enhance protein stability while retaining function. In adapting this method for a more limited number of protein sequences, we studied 21 consensus single-point variants from eight publicly available CD3 binding sequences with high similarity but diverse biophysical and pharmacological properties. All single-point consensus variants retained CD3 binding and performed similarly in cell-based functional assays. Using Ridge regression analysis, we identified the variants and sequence positions with overall beneficial effects on developability attributes of the CD3 binders. A second round of sequence generation that combined these substitutions into a single molecule yielded a unique CD3 binder with globally optimized developability attributes. In this first application to therapeutic antibodies, adapted Consensus Protein Design was found to be highly beneficial within lead optimization, conserving resources and minimizing iterations. Future implementations of this general strategy may help accelerate drug discovery and improve success rates in bringing novel biotherapeutics to market.
Keywords: Consensus protein design; bioinformatics; biotherapeutics; data science; developability; molecular modeling.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
