Functional and multiscale 3D structural investigation of brain tissue through correlative in vivo physiology, synchrotron microtomography and volume electron microscopy
- PMID: 35614048
- PMCID: PMC9132960
- DOI: 10.1038/s41467-022-30199-6
Functional and multiscale 3D structural investigation of brain tissue through correlative in vivo physiology, synchrotron microtomography and volume electron microscopy
Abstract
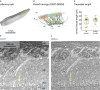
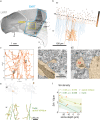
Understanding the function of biological tissues requires a coordinated study of physiology and structure, exploring volumes that contain complete functional units at a detail that resolves the relevant features. Here, we introduce an approach to address this challenge: Mouse brain tissue sections containing a region where function was recorded using in vivo 2-photon calcium imaging were stained, dehydrated, resin-embedded and imaged with synchrotron X-ray computed tomography with propagation-based phase contrast (SXRT). SXRT provided context at subcellular detail, and could be followed by targeted acquisition of multiple volumes using serial block-face electron microscopy (SBEM). In the olfactory bulb, combining SXRT and SBEM enabled disambiguation of in vivo-assigned regions of interest. In the hippocampus, we found that superficial pyramidal neurons in CA1a displayed a larger density of spine apparati than deeper ones. Altogether, this approach can enable a functional and structural investigation of subcellular features in the context of cells and tissues.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Young, B., Woodford, P. & O’Dowd, G. Wheater’s Functional Histology: a Text and Colour Atlas. 6th edn (Churchill Livingstone, 2000).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

