Molecular mechanism of the wake-promoting agent TAK-925
- PMID: 35614071
- PMCID: PMC9133036
- DOI: 10.1038/s41467-022-30601-3
Molecular mechanism of the wake-promoting agent TAK-925
Abstract
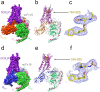
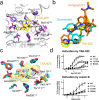
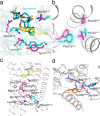
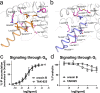
The OX2 orexin receptor (OX2R) is a highly expressed G protein-coupled receptor (GPCR) in the brain that regulates wakefulness and circadian rhythms in humans. Antagonism of OX2R is a proven therapeutic strategy for insomnia drugs, and agonism of OX2R is a potentially powerful approach for narcolepsy type 1, which is characterized by the death of orexinergic neurons. Until recently, agonism of OX2R had been considered 'undruggable.' We harness cryo-electron microscopy of OX2R-G protein complexes to determine how the first clinically tested OX2R agonist TAK-925 can activate OX2R in a highly selective manner. Two structures of TAK-925-bound OX2R with either a Gq mimetic or Gi reveal that TAK-925 binds at the same site occupied by antagonists, yet interacts with the transmembrane helices to trigger activating microswitches. Our structural and mutagenesis data show that TAK-925's selectivity is mediated by subtle differences between OX1 and OX2 receptor subtypes at the orthosteric pocket. Finally, differences in the polarity of interactions at the G protein binding interfaces help to rationalize OX2R's coupling selectivity for Gq signaling. The mechanisms of TAK-925's binding, activation, and selectivity presented herein will aid in understanding the efficacy of small molecule OX2R agonists for narcolepsy and other circadian disorders.
© 2022. The Author(s).
Conflict of interest statement
J.Y., K.C., R.R., P.L., X.B., J.K.D.B., and D.M.R. declare no competing interests. Y.K., A.P.M., M.S., E.K., A.O., T.K., Y.M., Y.S. and M.F. are current or former employees of Takeda Pharmaceutical Company Limited.
Figures

Comment in
-
Reply to: The G protein preference of orexin receptors is currently an unresolved issue.Nat Commun. 2023 Jun 1;14(1):3163. doi: 10.1038/s41467-023-38765-2. Nat Commun. 2023. PMID: 37264001 Free PMC article. No abstract available.
-
The G protein preference of orexin receptors is currently an unresolved issue.Nat Commun. 2023 Jun 1;14(1):3162. doi: 10.1038/s41467-023-38764-3. Nat Commun. 2023. PMID: 37264034 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

