Plin2-mediated lipid droplet mobilization accelerates exit from pluripotency by lipidomic remodeling and histone acetylation
- PMID: 35614132
- PMCID: PMC9613632
- DOI: 10.1038/s41418-022-01018-8
Plin2-mediated lipid droplet mobilization accelerates exit from pluripotency by lipidomic remodeling and histone acetylation
Abstract
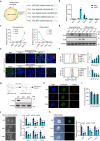
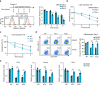
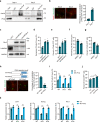
Metabolic switch is critical for cell fate determination through metabolic functions, epigenetic modifications, and gene expression. However, the mechanisms underlying these alterations and their functional roles remain unclear. Here, we show that Plin2-mediated moderate lipid hydrolysis is critical for pluripotency of embryonic stem cells (ESCs). Upon exit from pluripotency, lipid droplet (LD)-associated protein Plin2 is recognized by Hsc70 and degraded via chaperone-mediated autophagy to facilitate LD mobilization. Enhancing lipid hydrolysis by Plin2 knockout promotes pluripotency exit, which is recovered by ATGL inhibition. Mechanistically, excessive lipid hydrolysis induces a dramatic lipidomic remodeling characterized by decreased cardiolipin and phosphatidylethanolamine, which triggers defects in mitochondrial cristae and fatty acid oxidation, resulting in reduced acetyl-CoA and histone acetylation. Our results reveal how LD mobilization is regulated and its critical role in ESC pluripotency, and indicate the mechanism linking LD homeostasis to mitochondrial remodeling and epigenetic regulation, which might shed light on development and diseases.
© 2022. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

