CCR5 closes the temporal window for memory linking
- PMID: 35614219
- PMCID: PMC9197199
- DOI: 10.1038/s41586-022-04783-1
CCR5 closes the temporal window for memory linking
Abstract
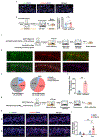
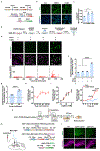
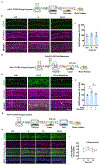
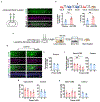
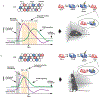
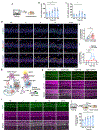
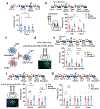
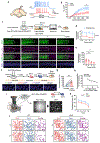
Real-world memories are formed in a particular context and are often not acquired or recalled in isolation1-5. Time is a key variable in the organization of memories, as events that are experienced close in time are more likely to be meaningfully associated, whereas those that are experienced with a longer interval are not1-4. How the brain segregates events that are temporally distinct is unclear. Here we show that a delayed (12-24 h) increase in the expression of C-C chemokine receptor type 5 (CCR5)-an immune receptor that is well known as a co-receptor for HIV infection6,7-after the formation of a contextual memory determines the duration of the temporal window for associating or linking that memory with subsequent memories. This delayed expression of CCR5 in mouse dorsal CA1 neurons results in a decrease in neuronal excitability, which in turn negatively regulates neuronal memory allocation, thus reducing the overlap between dorsal CA1 memory ensembles. Lowering this overlap affects the ability of one memory to trigger the recall of the other, and therefore closes the temporal window for memory linking. Our findings also show that an age-related increase in the neuronal expression of CCR5 and its ligand CCL5 leads to impairments in memory linking in aged mice, which could be reversed with a Ccr5 knockout and a drug approved by the US Food and Drug Administration (FDA) that inhibits this receptor, a result with clinical implications. Altogether, the findings reported here provide insights into the molecular and cellular mechanisms that shape the temporal window for memory linking.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures






Comment in
-
An immune molecule segregates memories in time.Nature. 2022 Jun;606(7912):38-39. doi: 10.1038/d41586-022-01166-4. Nature. 2022. PMID: 35614256 No abstract available.
-
Segregating memories: targeting microenvironment of neuronal ensembles.Signal Transduct Target Ther. 2022 Oct 12;7(1):363. doi: 10.1038/s41392-022-01214-5. Signal Transduct Target Ther. 2022. PMID: 36224166 Free PMC article. No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

