Transcriptome Analysis of Antennal Chemosensory Genes in Curculio Dieckmanni Faust. (Coleoptera: Curculionidae)
- PMID: 35615683
- PMCID: PMC9124802
- DOI: 10.3389/fphys.2022.896793
Transcriptome Analysis of Antennal Chemosensory Genes in Curculio Dieckmanni Faust. (Coleoptera: Curculionidae)
Abstract
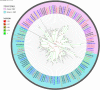
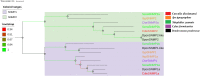
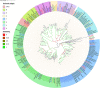
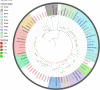
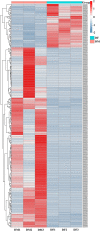
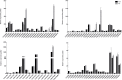
The olfactory system plays a key role in regulating insect behaviors, such as locating host plants, spawning sites, and mating partners and avoiding predators. Chemosensory genes are required for olfactory recognition in insects. Curculio dieckmanni Faust. (Coleoptera: Curculionidae) damages hazelnuts and causes severe economic losses. There are no effective control measures, but understanding the olfaction mechanisms of this insect could lead to a new approach for population management. However, the genes that perform chemosensory functions in C. dieckmanni are still unclear. Using high-throughput sequencing, we assembled the antennal transcriptome of C. dieckmanni and annotated the major chemosensory gene families. Of the chemosensory gene families, we found 23 odorant-binding proteins, 15 chemosensory proteins, 2 sensory neuron membrane proteins, 15 odorant receptors, 23 ionotropic receptors, and nine gustatory receptors. Using Blast sequence alignment and phylogenetic analysis, the sequences of these proteins were identified. Male- and female-specific chemosensory genes involved in odorant detection and recognition were validated by qRT-PCR. Among the chemosensory genes, we found significant differences in the expression of CdieOBP8, CdieOBP9, CdieOBP19, CdieOBP20, CdieOBP21, CdieCSP15, CdieOR13, and CdieOR15 between adult male and female C. dieckmanni. A total of 87 expressed chemosensory proteins were found in C. dieckmanni. Investigating these proteins will help reveal the molecular mechanism of odorant recognition in C. dieckmanni and may aid the development of novel control strategies for this species.
Keywords: antennal transcriptome; chemosensory genes; control strategies; curculio dieckmanni; curculionoidea.
Copyright © 2022 Ma, Lu, Zhang, Deng, Bai, Xu, Diao, Pang, Wang, Zhao, Ma and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Andersson M. N., Grosse-Wilde E., Keeling C. I., Bengtsson J. M., Yuen M. M., Li M., et al. (2013). Antennal Transcriptome Analysis of the Chemosensory Gene Families in the Tree Killing Bark Beetles, Ips Typographus and Dendroctonus Ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genomics 14 (14), 198–2164. 10.1186/1471-2164-14-198 - DOI - PMC - PubMed
-
- Announcement of the State Forestry Administration of China (2013). National Forestry Dangerous Pests List. (No.4 of 2013). Available at: http://www.forestry.gov.cn/portal/main/s/72/content-580486.html .
LinkOut - more resources
Full Text Sources
Research Materials

