A Simple Standard for Sharing Ontological Mappings (SSSOM)
- PMID: 35616100
- PMCID: PMC9216545
- DOI: 10.1093/database/baac035
A Simple Standard for Sharing Ontological Mappings (SSSOM)
Abstract
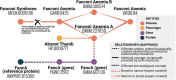
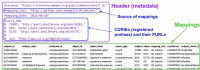
Despite progress in the development of standards for describing and exchanging scientific information, the lack of easy-to-use standards for mapping between different representations of the same or similar objects in different databases poses a major impediment to data integration and interoperability. Mappings often lack the metadata needed to be correctly interpreted and applied. For example, are two terms equivalent or merely related? Are they narrow or broad matches? Or are they associated in some other way? Such relationships between the mapped terms are often not documented, which leads to incorrect assumptions and makes them hard to use in scenarios that require a high degree of precision (such as diagnostics or risk prediction). Furthermore, the lack of descriptions of how mappings were done makes it hard to combine and reconcile mappings, particularly curated and automated ones. We have developed the Simple Standard for Sharing Ontological Mappings (SSSOM) which addresses these problems by: (i) Introducing a machine-readable and extensible vocabulary to describe metadata that makes imprecision, inaccuracy and incompleteness in mappings explicit. (ii) Defining an easy-to-use simple table-based format that can be integrated into existing data science pipelines without the need to parse or query ontologies, and that integrates seamlessly with Linked Data principles. (iii) Implementing open and community-driven collaborative workflows that are designed to evolve the standard continuously to address changing requirements and mapping practices. (iv) Providing reference tools and software libraries for working with the standard. In this paper, we present the SSSOM standard, describe several use cases in detail and survey some of the existing work on standardizing the exchange of mappings, with the goal of making mappings Findable, Accessible, Interoperable and Reusable (FAIR). The SSSOM specification can be found at http://w3id.org/sssom/spec. Database URL: http://w3id.org/sssom/spec.
© The Author(s) 2022. Published by Oxford University Press.
Figures

References
-
- Broeder D., Budroni P., Degl’Innocenti E.. et al. (2021) SEMAF: A proposal for a flexible semantic mapping framework. https://zenodo.org/record/4651421#.Yn60VBPMKkg.
-
- Laadhar A., Abrahão E. and Jonquet C. (2020) Investigating one million XRefs in thirthy ontologies from the OBO world. In: 11th International Conference on Biomedical Ontologies (ICBO).
-
- Alignment API . https://moex.gitlabpages.inria.fr/alignapi/ (20 November 2021, date last accessed).
-
- Jackson R., Matentzoglu N., Overton J.A.. et al. (2021) OBO Foundry in 2021: operationalizing open data principles to evaluate ontologies. Database, 2021. 10.1093/database/baab069. - DOI - PMC - PubMed
-
- Laadhar A., Abrahão E. and Jonquet C. (2020) Investigating one million XRefs in thirthy ontologies from the OBO world. In: ICBO 2020-11th International Conference on Biomedical Ontologies, Vol. 2807, pp. G.1–12.

