Clinical validity assessment of genes frequently tested on intellectual disability/autism sequencing panels
- PMID: 35616647
- PMCID: PMC10200330
- DOI: 10.1016/j.gim.2022.05.001
Clinical validity assessment of genes frequently tested on intellectual disability/autism sequencing panels
Abstract
Purpose: Neurodevelopmental disorders (NDDs), such as intellectual disability (ID) and autism spectrum disorder (ASD), exhibit genetic and phenotypic heterogeneity, making them difficult to differentiate without a molecular diagnosis. The Clinical Genome Resource Intellectual Disability/Autism Gene Curation Expert Panel (GCEP) uses systematic curation to distinguish ID/ASD genes that are appropriate for clinical testing (ie, with substantial evidence supporting their relationship to disease) from those that are not.
Methods: Using the Clinical Genome Resource gene-disease validity curation framework, the ID/Autism GCEP classified genes frequently included on clinical ID/ASD testing panels as Definitive, Strong, Moderate, Limited, Disputed, Refuted, or No Known Disease Relationship.
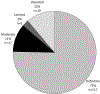
Results: As of September 2021, 156 gene-disease pairs have been evaluated. Although most (75%) were determined to have definitive roles in NDDs, 22 (14%) genes evaluated had either Limited or Disputed evidence. Such genes are currently not recommended for use in clinical testing owing to the limited ability to assess the effect of identified variants.
Conclusion: Our understanding of gene-disease relationships evolves over time; new relationships are discovered and previously-held conclusions may be questioned. Without periodic re-examination, inaccurate gene-disease claims may be perpetuated. The ID/Autism GCEP will continue to evaluate these claims to improve diagnosis and clinical care for NDDs.
Keywords: Autism; ClinGen; Gene–disease validity; Intellectual disability; Neurodevelopmental disorders.
Copyright © 2022 American College of Medical Genetics and Genomics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest A.Br. is a shareholder of and employed by Natera. A.Br. has also been an employee of Invitae and Quest Diagnostics commercial laboratories. A.R.C. and K.B. are shareholders of and employed by Illumina, Inc. A.Be. is a shareholder of and is employed by Invitae. B.B. has received research support from Biomarin Pharmaceuticals Inc. He is currently employed by and is a shareholder of Alnylam Pharmaceuticals, Inc. All other authors declare no conflicts of interest.
Figures
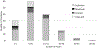
References
-
- Srivastava S, Love-Nichols JA, Dies KA, et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med. 2019;21(11):2413–2421. Published correction appears in Genet Med. 2020;22(10):1731–1732. 10.1038/s41436-019-0554-6 - DOI - PMC - PubMed

