Interaction of fungal lipase with potential phytotherapeutics
- PMID: 35617167
- PMCID: PMC9135303
- DOI: 10.1371/journal.pone.0264460
Interaction of fungal lipase with potential phytotherapeutics
Abstract
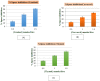
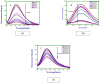
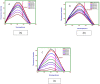
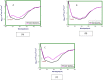
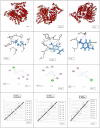
Interaction of thymol, carvacrol and linalool with fungal lipase and Human Serum Albumin (HSA) have been investigated employing UV-Vis spectroscopy Fluorescence and Circular dichroism spectroscopy (CD) along with docking studies. Thymol, carvacrol and linalool displayed approximately 50% inhibition at 1.5 mmol/litre concentrations using para-nitrophenyl palmitate (pNPP). UV-Vis spectroscopy give evidence of the formation of lipase-linalool, lipase-carvacrol and lipase-thymol complex at the ground state. Three molecules also showed complex formation with HSA at the ground state. Fluorescence spectroscopy shows strong binding of lipase to thymol (Ka of 2.6 x 109 M-1) as compared to carvacrol (4.66 x 107 M-1) and linalool (5.3 x 103 M-1). Number of binding sites showing stoichiometry of association process on lipase is found to be 2.52 (thymol) compared to 2.04 (carvacrol) and 1.12 (linalool). Secondary structure analysis by CD spectroscopy results, following 24 hours incubation at 25°C, with thymol, carvacrol and linalool revealed decrease in negative ellipticity for lipase indicating loss in helical structure as compared with the native protein. The lowering in negative ellipticity was in the order of thymol > carvacrol > linalool. Fluorescence spectra following binding of all three molecules with HSA caused blue shift which suggests the compaction of the HSA structure. Association constant of thymol and HSA is 9.6 x 108 M-1 which along with 'n' value of 2.41 suggests strong association and stable complex formation, association constant for carvacrol and linalool was in range of 107 and 103 respectively. Docking results give further insight into strong binding of thymol, carvacrol and linalool with lipase having free energy of binding as -7.1 kcal/mol, -5.0 kcal/mol and -5.2 kcal/mol respectively. To conclude, fungal lipases can be attractive target for controlling their growth and pathogenicity. Employing UV-Vis, Fluorescence and Circular dichroism spectroscopy we have shown that thymol, carvacrol and linalool strongly bind and disrupt structure of fungal lipase, these three phytochemicals also bind well with HSA. Based on disruption of lipase structure and its binding nature with HSA, we concluded thymol as a best anti-lipase molecule among three molecules tested. Results of Fluorescence and CD spectroscopy taken together suggests that thymol and carvacrol are profound disrupter of lipase structure.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Alviano W, Mendonça‐Filho R, Alviano D, Bizzo H, Souto‐Padrón T, Rodrigues M, et al.. Antimicrobial activity of Croton cajucara Benth linalool‐rich essential oil on artificial biofilms and planktonic microorganisms. Oral microbiology and immunology. 2005;20(2):101–5. doi: 10.1111/j.1399-302X.2004.00201.x - DOI - PubMed
-
- de Castro RD, de Souza TMPA, Bezerra LMD, Ferreira GLS, Costa EMMdB, Cavalcanti AL. Antifungal activity and mode of action of thymol and its synergism with nystatin against Candida species involved with infections in the oral cavity: an in vitro study. BMC complementary and alternative medicine. 2015;15:417-. doi: 10.1186/s12906-015-0947-2 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

