Integrated bioinformatics and statistical approaches to explore molecular biomarkers for breast cancer diagnosis, prognosis and therapies
- PMID: 35617355
- PMCID: PMC9135200
- DOI: 10.1371/journal.pone.0268967
Integrated bioinformatics and statistical approaches to explore molecular biomarkers for breast cancer diagnosis, prognosis and therapies
Abstract
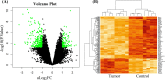
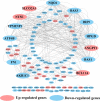
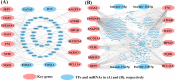
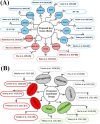
Integrated bioinformatics and statistical approaches are now playing the vital role in identifying potential molecular biomarkers more accurately in presence of huge number of alternatives for disease diagnosis, prognosis and therapies by reducing time and cost compared to the wet-lab based experimental procedures. Breast cancer (BC) is one of the leading causes of cancer related deaths for women worldwide. Several dry-lab and wet-lab based studies have identified different sets of molecular biomarkers for BC. But they did not compare their results to each other so much either computationally or experimentally. In this study, an attempt was made to propose a set of molecular biomarkers that might be more effective for BC diagnosis, prognosis and therapies, by using the integrated bioinformatics and statistical approaches. At first, we identified 190 differentially expressed genes (DEGs) between BC and control samples by using the statistical LIMMA approach. Then we identified 13 DEGs (AKR1C1, IRF9, OAS1, OAS3, SLCO2A1, NT5E, NQO1, ANGPT1, FN1, ATF6B, HPGD, BCL11A, and TP53INP1) as the key genes (KGs) by protein-protein interaction (PPI) network analysis. Then we investigated the pathogenetic processes of DEGs highlighting KGs by GO terms and KEGG pathway enrichment analysis. Moreover, we disclosed the transcriptional and post-transcriptional regulatory factors of KGs by their interaction network analysis with the transcription factors (TFs) and micro-RNAs. Both supervised and unsupervised learning's including multivariate survival analysis results confirmed the strong prognostic power of the proposed KGs. Finally, we suggested KGs-guided computationally more effective seven candidate drugs (NVP-BHG712, Nilotinib, GSK2126458, YM201636, TG-02, CX-5461, AP-24534) compared to other published drugs by cross-validation with the state-of-the-art alternatives top-ranked independent receptor proteins. Thus, our findings might be played a vital role in breast cancer diagnosis, prognosis and therapies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Koo MM, von Wagner C, Abel GA, McPhail S, Rubin GP, Lyratzopoulos G. Typical and atypical presenting symptoms of breast cancer and their associations with diagnostic intervals: Evidence from a national audit of cancer diagnosis. Cancer Epidemiol. 2017; 48:140–6. doi: 10.1016/j.canep.2017.04.010 . - DOI - PMC - PubMed
-
- Cancer.Net. Breast Cancer: Statistics 2021. Available from: https://www.cancer.net/cancer-types/breast-cancer/statistics.
-
- Mosharaf MP, Reza MS, Kibria MK, Ahmed FF, Kabir MH, Hasan S, et al.. Computational identification of host genomic biomarkers highlighting their functions, pathways and regulators that influence SARS-CoV-2 infections and drug repurposing. Sci Rep. 2022; 12(1):4279. doi: 10.1038/s41598-022-08073-8 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

