Best practice standards for circular RNA research
- PMID: 35618955
- PMCID: PMC9759028
- DOI: 10.1038/s41592-022-01487-2
Best practice standards for circular RNA research
Abstract
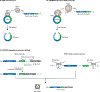
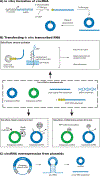
Circular RNAs (circRNAs) are formed in all domains of life and via different mechanisms. There has been an explosion in the number of circRNA papers in recent years; however, as a relatively young field, circRNA biology has an urgent need for common experimental standards for isolating, analyzing, expressing and depleting circRNAs. Here we propose a set of guidelines for circRNA studies based on the authors' experience. This Perspective will specifically address the major class of circRNAs in Eukarya that are generated by a spliceosome-catalyzed back-splicing event. We hope that the implementation of best practice principles for circRNA research will help move the field forward and allow a better functional understanding of this fascinating group of RNAs.
© 2022. Springer Nature America, Inc.
Conflict of interest statement
Competing Interests statement:
The authors have no competing interests
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

