Transcriptome-Wide Association Studies and Integration Analysis of mRNA Expression Profiles Identify Candidate Genes and Pathways Associated With Ankylosing Spondylitis
- PMID: 35619696
- PMCID: PMC9128383
- DOI: 10.3389/fimmu.2022.814303
Transcriptome-Wide Association Studies and Integration Analysis of mRNA Expression Profiles Identify Candidate Genes and Pathways Associated With Ankylosing Spondylitis
Abstract
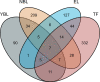
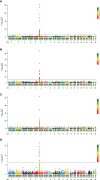
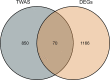
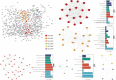
This study aimed to identify susceptibility genes and pathways associated with ankylosing spondylitis (AS) by integrating whole transcriptome-wide association study (TWAS) analysis and mRNA expression profiling data. AS genome-wide association study (GWAS) summary data from the large GWAS database were used. This included data of 1265 AS patients and 452264 controls. A TWAS of AS was conducted using these data. The analysis software used was FUSION, and Epstein-Barr virus-transformed lymphocytes, transformed fibroblasts, peripheral blood, and whole blood were used as gene expression references. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed for the important genes identified via TWAS. Protein-protein interaction (PPI) network analysis based on the STRING database was also performed to detect genes shared by TWAS and mRNA expression profiles in AS. TWAS identified 920 genes (P <0.05) and analyzed mRNA expression profiles to obtain 1183 differential genes. Following comparison of the TWAS results and mRNA expression characteristics, we obtained 70 overlapping genes and performed GO and KEGG enrichment analyses of these genes to obtain 16 pathways. Via PPI network analysis, we obtained the protein interaction network and performed MCODE analysis to acquire the HUB genes. Similarly, we performed GO and KEGG analyses on the genes identified by TWAS, obtained 98 pathways after screening, and analyzed protein interactions via the PPI network. Through the integration of TWAS and mRNA expression analysis, genes related to AS and GO and KEGG terms were determined, providing new evidence and revealing the pathogenesis of AS. Our AS TWAS work identified novel genes associated with AS, as well as suggested potential tissues and pathways of action for these TWAS AS genes, providing a new direction for research into the pathogenesis of AS.
Keywords: ankylosing spondylitis; genome-wide association study (GWAS); mRNA expression profile; susceptibility genes; transcriptome-wide association study (TWAS).
Copyright © 2022 Feng, Lu, Liu, Xu and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Arends S, Spoorenberg A, Bruyn GA, Houtman PM, Leijsma MK, Kallenberg CG, et al. . The Relation Between Bone Mineral Density, Bone Turnover Markers, and Vitamin D Status in Ankylosing Spondylitis Patients With Active Disease: A Cross-Sectional Analysis. Osteoporosis Int J Established As Result Cooperation Between Eur Foundation Osteoporosis Natl Osteoporosis Foundation USA (2011) 22(5):1431–9. doi: 10.1007/s00198-010-1338-7 - DOI - PMC - PubMed
-
- de Winter JJ, van Mens LJ, van der Heijde D, Landewé R, Baeten DL. Prevalence of Peripheral and Extra-Articular Disease in Ankylosing Spondylitis Versus Non-Radiographic Axial Spondyloarthritis: A Meta-Analysis. Arthritis Res Ther (2016) 18(1):196–6. doi: 10.1186/s13075-016-1093-z - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

