Landscape of Immune Cells Heterogeneity in Liver Transplantation by Single-Cell RNA Sequencing Analysis
- PMID: 35619708
- PMCID: PMC9127089
- DOI: 10.3389/fimmu.2022.890019
Landscape of Immune Cells Heterogeneity in Liver Transplantation by Single-Cell RNA Sequencing Analysis
Abstract
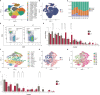
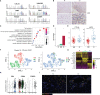
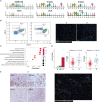
Rejection is still a critical barrier to the long-term survival of graft after liver transplantation, requiring clinicians to unveil the underlying mechanism of liver transplant rejection. The cellular diversity and the interplay between immune cells in the liver graft microenvironment remain unclear. Herein, we performed single-cell RNA sequencing analysis to delineate the landscape of immune cells heterogeneity in liver transplantation. T cells, NK cells, B cells, and myeloid cell subsets in human liver and blood were enriched to characterize their tissue distribution, gene expression, and functional modules. The proportion of CCR6+CD4+ T cells increased within an allograft, suggesting that there are more memory CD4+ T cells after transplantation, in parallel with exhausted CTLA4+CD8+ T and actively proliferating MKI67+CD8+ T cells increased significantly, where they manifested heterogeneity, distinct function, and homeostatic proliferation. Remarkably, the changes of CD1c+ DC, CADM+ DC, MDSC, and FOLR3+ Kupffer cells increase significantly, but the proportion of CD163+ Kupffer, APOE+ Kupffer, and GZMA+ Kupffer decreased. Furthermore, we identified LDLR as a novel marker of activated MDSC to prevent liver transplant rejection. Intriguingly, a subset of CD4+CD8+FOXP3+ T cells included in CTLA4+CD8+ T cells was first detected in human liver transplantation. Furthermore, intercellular communication and gene regulatory analysis implicated the LDLR+ MDSC and CTLA4+CD8+ T cells interact through TIGIT-NECTIN2 signaling pathway. Taken together, these findings have gained novel mechanistic insights for understanding the immune landscape in liver transplantation, and it outlines the characteristics of immune cells and provides potential therapeutic targets in liver transplant rejection.
Keywords: CD8 T cell; MDSC; immune; liver tranpslant; single cell RNA sequence.
Copyright © 2022 Li, Li, Wu, Xu, Teng, Yang, Sun, Zhao, Li, Liu, Yang, Gong and Cai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

