Detoxification of Aflatoxin B1 by a Potential Probiotic Bacillus amyloliquefaciens WF2020
- PMID: 35620100
- PMCID: PMC9127598
- DOI: 10.3389/fmicb.2022.891091
Detoxification of Aflatoxin B1 by a Potential Probiotic Bacillus amyloliquefaciens WF2020
Abstract
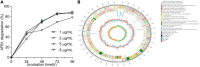
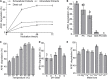
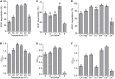
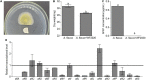
Microbial degradation is considered as an attractive method to eliminate exposure to aflatoxin B1 (AFB1), the most toxic mycotoxin that causes great economic losses and brings a serious threat to human and animal health, in food and feed. In this study, Bacillus amyloliquefaciens WF2020, isolated from naturally fermented pickles, could effectively degrade AFB1 ranging from 1 to 8 μg/ml, and the optimum temperature and pH value were 37-45°C and 8.0, respectively. Moreover, B. amyloliquefaciens WF2020 was considered to be a potential probiotic due to the synthesis of active compounds, absence of virulence genes, susceptibility to various antibiotics, and enhanced lifespan of Caenorhabditis elegans. Extracellular enzymes or proteins played a major role in AFB1 degradation mediated by B. amyloliquefaciens WF2020 into metabolites with low or no mutagenicity and toxicity to C. elegans. AFB1 degradation by the cell-free supernatant was stable up to 70°C, with an optimal pH of 8.0, and the cell-free supernatant could still degrade AFB1 by 37.16% after boiling for 20 min. Furthermore, B. amyloliquefaciens WF2020 caused a slight defect in fungal growth and completely inhibited AFB1 production when co-incubated with Aspergillus flavus. Additionally, B. amyloliquefaciens WF2020 suppressed the expression of 10 aflatoxin pathway genes and 2 transcription factors (alfR and alfS), suggesting that B. amyloliquefaciens WF2020 might inhibit AFB1 synthesis in A. flavus. These results indicate that B. amyloliquefaciens WF2020 and/or its extracellular enzymes or proteins have a promising potential to be applied in protecting food and feed from AFB1 contamination.
Keywords: Ames test; Aspergillus flavus; Bacillus amyloliquefaciens; Caenorhabditis elegans; aflatoxin B1; genome sequence.
Copyright © 2022 Chen, Fang, Liao, Xu, Liang, Liu, Zhong, Wang, Fang and Wang.
Conflict of interest statement
CX and ZhiL are employed by Guangdong Moyanghua Grains and Oils Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Abdel-Shafi S., Shehata S., Shindia A., Ei-Meligy K., Khidr A. (2018). Biodegradation of aflatoxins by bacteria. Egyptian J. Microbiol. 53 241–254. 10.21608/ejm.2018.5752.1078 - DOI
LinkOut - more resources
Full Text Sources

