T-cell activation and bacterial infection in skin wounds of recessive dystrophic epidermolysis bullosa patients
- PMID: 35620886
- PMCID: PMC9541540
- DOI: 10.1111/exd.14615
T-cell activation and bacterial infection in skin wounds of recessive dystrophic epidermolysis bullosa patients
Abstract
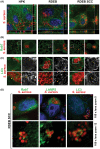
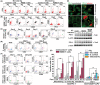
Recessive dystrophic epidermolysis bullosa (RDEB) patients develop poorly healing skin wounds that are frequently colonized with microbiota. Because T cells play an important role in clearing such pathogens, we aimed to define the status of adaptive T cell-mediated immunity in RDEB wounds. Using a non-invasive approach for sampling of wound-associated constituents, we evaluated microbial contaminants in cellular fraction and exudates obtained from RDED wounds. Infectivity and intracellular trafficking of inactivated Staphylococcus aureus was accessed in RDEB keratinocytes. S. aureus and microbial antigen-specific activation of RDEB wound-derived T cells were investigated by fluorescence-activated cell sorting-based immune-phenotyping and T-cell functional assays. We found that RDEB wounds and epithelial cells are most frequently infected with Staphylococcus sp. and Pseudomonas sp. and that S. aureus essentially infects more RDEB keratinocytes and RDEB-derived squamous cell carcinoma cells than keratinocytes from healthy donors. The RDEB wound-associated T cells contain populations of CD4+ and CD8+ peripheral memory T cells that respond to soluble microbial antigens by proliferating and secreting interferon gamma (IFNγ). Moreover, CD8+ cytotoxic T lymphocytes recognize S. aureus-infected RDEB keratinocytes and respond by producing interleukin-2 (IL-2) and IFNγ and degranulating and cytotoxically killing infected cells. Prolonged exposure of RDEB-derived T cells to microbial antigens in vitro does not trigger PD-1-mediated T-cell exhaustion but induces differentiation of the CD4high population into CD4high CD25+ FoxP3+ regulatory T cells. Our data demonstrated that adaptive T cell-mediated immunity could clear infected cells from wound sites, but these effects might be inhibited by PD-1/Treg-mediated immuno-suppression in RDEB.
Keywords: RDEB; T-cell immunity; infection; wound healing.
© 2022 The Authors. Experimental Dermatology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Has C, Nystrom A, Saeidian AH, Bruckner‐Tuderman L, Uitto J. Epidermolysis bullosa: molecular pathology of connective tissue components in the cutaneous basement membrane zone. Matrix Biol. 2018;71‐72:313‐329. - PubMed
-
- Mellerio J.E., Infection and colonization in epidermolysis bullosa, Dermatol Clin. 28(2) (2010) 267–9, ix. - PubMed
-
- Phillips T, Huitema L, Cepeda R, et al. Aberrant recruitment of leukocytes defines poor wound healing in patients with recessive dystrophic epidermolysis bullosa. J Dermatol Sci. 2020;100(3):209‐216. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

