Temporal Expression Profiles Reveal Potential Targets during Postembryonic Development of Forensically Important Sarcophaga peregrina (Diptera: Sarcophagidae)
- PMID: 35621788
- PMCID: PMC9143129
- DOI: 10.3390/insects13050453
Temporal Expression Profiles Reveal Potential Targets during Postembryonic Development of Forensically Important Sarcophaga peregrina (Diptera: Sarcophagidae)
Abstract
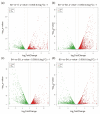
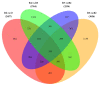
Sarcophaga peregrina (Robineau-Desvoidy, 1830) is a species of medical and forensic importance. In order to investigate the molecular mechanism during postembryonic development and identify specific genes that may serve as potential targets, transcriptome analysis was used to investigate its gene expression dynamics from the larval to pupal stages, based on our previous de novo-assembled genome of S. peregrina. Totals of 2457, 3656, 3764, and 2554 differentially expressed genes were identified. The specific genes encoding the structural constituent of cuticle were significantly differentially expressed, suggesting that degradation and synthesis of cuticle-related proteins might actively occur during metamorphosis. Molting (20-hydroxyecdysone, 20E) and juvenile (JH) hormone pathways were significantly enriched, and gene expression levels changed in a dynamic pattern during the developmental stages. In addition, the genes in the oxidative phosphorylation pathway were significantly expressed at a high level during the larval stage, and down-regulated from the wandering to pupal stages. Weighted gene co-expression correlation network analysis (WGCNA) further demonstrated the potential regulation mechanism of tyrosine metabolism in the process of puparium tanning. Moreover, 10 consistently up-regulated genes were further validated by qRT-PCR. The utility of the models was then examined in a blind study, indicating the ability to predict larval development. The developmental, stage-specific gene profiles suggest novel molecular markers for age prediction of forensically important flies.
Keywords: Sarcophaga peregrina; age estimation; gene expression; metamorphosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
Grants and funding
LinkOut - more resources
Full Text Sources

