Metabolic signatures of pregnancy-induced cardiac growth
- PMID: 35622533
- PMCID: PMC9236881
- DOI: 10.1152/ajpheart.00105.2022
Metabolic signatures of pregnancy-induced cardiac growth
Abstract
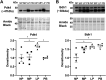
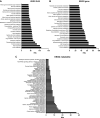
The goal of this study was to develop an atlas of the metabolic, transcriptional, and proteomic changes that occur with pregnancy in the maternal heart. Timed pregnancy studies in FVB/NJ mice revealed a significant increase in heart size by day 8 of pregnancy (midpregnancy; MP), which was sustained throughout the rest of the term compared with nonpregnant control mice. Cardiac hypertrophy and myocyte cross-sectional area were highest 7 days after birth (postbirth; PB) and were associated with significant increases in end-diastolic and end-systolic left ventricular volumes and higher cardiac output. Metabolomics analyses revealed that by day 16 of pregnancy (late pregnancy; LP) metabolites associated with nitric oxide production as well as acylcholines, sphingomyelins, and fatty acid species were elevated, which coincided with a lower activation state of phosphofructokinase and higher levels of pyruvate dehydrogenase kinase 4 (Pdk4) and β-hydroxybutyrate dehydrogenase 1 (Bdh1). In the postpartum period, urea cycle metabolites, polyamines, and phospholipid levels were markedly elevated in the maternal heart. Cardiac transcriptomics in LP revealed significant increases in not only Pdk4 and Bdh1 but also genes that regulate glutamate and ketone body oxidation, which were preceded in MP by higher expression of transcripts controlling cell proliferation and angiogenesis. Proteomics analysis of the maternal heart in LP and PB revealed significant reductions in several contractile filament and mitochondrial subunit complex proteins. Collectively, these findings describe the coordinated molecular changes that occur in the maternal heart during and after pregnancy.NEW & NOTEWORTHY Little is known of the underlying molecular and cellular mechanisms that contribute to pregnancy-induced cardiac growth. Several lines of evidence suggest that changes in cardiac metabolism may contribute. Here, we provide a comprehensive metabolic atlas of the metabolomic, proteomic, and transcriptomic changes occurring in the maternal heart. We show that pregnancy-induced cardiac growth is associated with changes in glycerophospholipid, nucleotide, and amino acid metabolism, with reductions in cardiac glucose catabolism. Collectively, these results suggest that substantial metabolic changes occur in the maternal heart during and after pregnancy.
Keywords: hypertrophy; metabolomics; pregnancy; proteomics; transcriptomics.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures




References
Publication types
MeSH terms
Associated data
Grants and funding
- HL154663/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- GM103436/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- GM127607/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- P30 GM127607/GM/NIGMS NIH HHS/United States
- R24 GM137786/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources

