Discovering sparse control strategies in neural activity
- PMID: 35622828
- PMCID: PMC9140285
- DOI: 10.1371/journal.pcbi.1010072
Discovering sparse control strategies in neural activity
Abstract
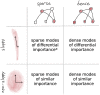
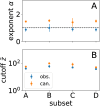
Biological circuits such as neural or gene regulation networks use internal states to map sensory input to an adaptive repertoire of behavior. Characterizing this mapping is a major challenge for systems biology. Though experiments that probe internal states are developing rapidly, organismal complexity presents a fundamental obstacle given the many possible ways internal states could map to behavior. Using C. elegans as an example, we propose a protocol for systematic perturbation of neural states that limits experimental complexity and could eventually help characterize collective aspects of the neural-behavioral map. We consider experimentally motivated small perturbations-ones that are most likely to preserve natural dynamics and are closer to internal control mechanisms-to neural states and their impact on collective neural activity. Then, we connect such perturbations to the local information geometry of collective statistics, which can be fully characterized using pairwise perturbations. Applying the protocol to a minimal model of C. elegans neural activity, we find that collective neural statistics are most sensitive to a few principal perturbative modes. Dominant eigenvalues decay initially as a power law, unveiling a hierarchy that arises from variation in individual neural activity and pairwise interactions. Highest-ranking modes tend to be dominated by a few, "pivotal" neurons that account for most of the system's sensitivity, suggesting a sparse mechanism of collective control.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

