Effector loss drives adaptation of Pseudomonas syringae pv. actinidiae biovar 3 to Actinidia arguta
- PMID: 35622878
- PMCID: PMC9182610
- DOI: 10.1371/journal.ppat.1010542
Effector loss drives adaptation of Pseudomonas syringae pv. actinidiae biovar 3 to Actinidia arguta
Abstract
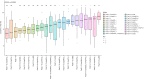
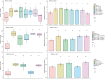
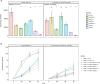
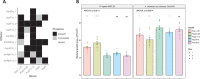
A pandemic isolate of Pseudomonas syringae pv. actinidiae biovar 3 (Psa3) has devastated kiwifruit orchards growing cultivars of Actinidia chinensis. In contrast, A. arguta (kiwiberry) is not a host of Psa3. Resistance is mediated via effector-triggered immunity, as demonstrated by induction of the hypersensitive response in infected A. arguta leaves, observed by microscopy and quantified by ion-leakage assays. Isolates of Psa3 that cause disease in A. arguta have been isolated and analyzed, revealing a 51 kb deletion in the exchangeable effector locus (EEL). This natural EEL-mutant isolate and strains with synthetic knockouts of the EEL were more virulent in A. arguta plantlets than wild-type Psa3. Screening of a complete library of Psa3 effector knockout strains identified increased growth in planta for knockouts of four effectors-AvrRpm1a, HopF1c, HopZ5a, and the EEL effector HopAW1a -suggesting a resistance response in A. arguta. Hypersensitive response (HR) assays indicate that three of these effectors trigger a host species-specific HR. A Psa3 strain with all four effectors knocked out escaped host recognition, but a cumulative increase in bacterial pathogenicity and virulence was not observed. These avirulence effectors can be used in turn to identify the first cognate resistance genes in Actinidia for breeding durable resistance into future kiwifruit cultivars.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Bull CT, De Boer S, Denny T, Firrao G, Saux MF-L, Saddler G, et al.. Comprehensive list of names of plant pathogenic bacteria, 1980–2007. J Plant Pathol. 2010:551–92.
-
- Morris CE, Lamichhane JR, Nikolić I, Stanković S, Moury B. The overlapping continuum of host range among strains in the Pseudomonas syringae complex. Phytopathology Research. 2019;1(1):4.
-
- Serizawa S, Ichikawa T, Takikawa Y, Tsuyumu S, Goto M. Occurrence of bacterial canker of kiwifruit in Japan. Japanese Journal of Phytopathology. 1989;55(4):427–36.
-
- Takikawa Y, Serizawa S, Ichikawa T, Tsuyumu S, Goto M. Pseudomonas syringae pv. actinidiae pv. nov. the causal bacterium of canker of kiwifruit in Japan. Japanese Journal of Phytopathology. 1989;55(4):437–44.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

