Production of 3',3'-cGAMP by a Bdellovibrio bacteriovorus promiscuous GGDEF enzyme, Bd0367, regulates exit from prey by gliding motility
- PMID: 35622882
- PMCID: PMC9140294
- DOI: 10.1371/journal.pgen.1010164
Production of 3',3'-cGAMP by a Bdellovibrio bacteriovorus promiscuous GGDEF enzyme, Bd0367, regulates exit from prey by gliding motility
Abstract
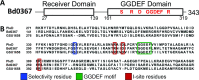
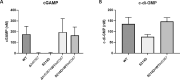
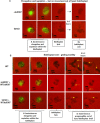
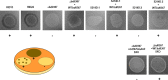
Bacterial second messengers are important for regulating diverse bacterial lifestyles. Cyclic di-GMP (c-di-GMP) is produced by diguanylate cyclase enzymes, named GGDEF proteins, which are widespread across bacteria. Recently, hybrid promiscuous (Hypr) GGDEF proteins have been described in some bacteria, which produce both c-di-GMP and a more recently identified bacterial second messenger, 3',3'-cyclic-GMP-AMP (cGAMP). One of these proteins was found in the predatory Bdellovibrio bacteriovorus, Bd0367. The bd0367 GGDEF gene deletion strain was found to enter prey cells, but was incapable of leaving exhausted prey remnants via gliding motility on a solid surface once predator cell division was complete. However, it was unclear which signal regulated this process. We show that cGAMP signalling is active within B. bacteriovorus and that, in addition to producing c-di-GMP and some c-di-AMP, Bd0367 is a primary producer of cGAMP in vivo. Site-directed mutagenesis of serine 214 to an aspartate rendered Bd0367 into primarily a c-di-GMP synthase. B. bacteriovorus strain bd0367S214D phenocopies the bd0367 deletion strain by being unable to glide on a solid surface, leading to an inability of new progeny to exit from prey cells post-replication. Thus, this process is regulated by cGAMP. Deletion of bd0367 was also found to be incompatible with wild-type flagellar biogenesis, as a result of an acquired mutation in flagellin chaperone gene homologue fliS, implicating c-di-GMP in regulation of swimming motility. Thus the single Bd0367 enzyme produces two secondary messengers by action of the same GGDEF domain, the first reported example of a synthase that regulates multiple second messengers in vivo. Unlike roles of these signalling molecules in other bacteria, these signal to two separate motility systems, gliding and flagellar, which are essential for completion of the bacterial predation cycle and prey exit by B. bacteriovorus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

