Spatiotemporal co-dependency between macrophages and exhausted CD8+ T cells in cancer
- PMID: 35623342
- PMCID: PMC9197962
- DOI: 10.1016/j.ccell.2022.05.004
Spatiotemporal co-dependency between macrophages and exhausted CD8+ T cells in cancer
Abstract
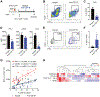
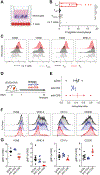
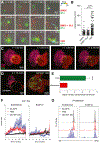
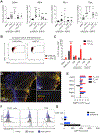
T cell exhaustion is a major impediment to antitumor immunity. However, it remains elusive how other immune cells in the tumor microenvironment (TME) contribute to this dysfunctional state. Here, we show that the biology of tumor-associated macrophages (TAMs) and exhausted T cells (Tex) in the TME is extensively linked. We demonstrate that in vivo depletion of TAMs reduces exhaustion programs in tumor-infiltrating CD8+ T cells and reinvigorates their effector potential. Reciprocally, transcriptional and epigenetic profiling reveals that Tex express factors that actively recruit monocytes to the TME and shape their differentiation. Using lattice light sheet microscopy, we show that TAM and CD8+ T cells engage in unique, long-lasting, antigen-specific synaptic interactions that fail to activate T cells but prime them for exhaustion, which is then accelerated in hypoxic conditions. Spatially resolved sequencing supports a spatiotemporal self-enforcing positive feedback circuit that is aligned to protect rather than destroy a tumor.
Keywords: T cell exhaustion; antitumor immunity; immunotherapy; tumor microenvironment; tumor-associated macrophages.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.T.S. is a scientific founder of Immunai and founder of Cartography Biosciences and receives research funding from Merck Research Laboratories and Allogene Therapeutics. M.F.K. is a founder and shareholder in Pionyr Immunotherapeutics and in Foundery Innovations, which prosecute and develop novel immunotherapeutics, respectively. The other authors declare no competing interests.
Figures


References
-
- Argüello RJ, Combes AJ, Char R, Gigan J-P, Baaziz AI, Bousiquot E, Camosseto V, Samad B, Tsui J, Yan P, et al. (2020). SCENITH: A Flow Cytometry-Based Method to Functionally Profile Energy Metabolism with Single-Cell Resolution. Cell Metab. 32, 1063–1075. 10.1016/j.cmet.2020.11.007. - DOI - PMC - PubMed
-
- Beltra J-C, Manne S, Abdel-Hakeem MS, Kurachi M, Giles JR, Chen Z, Casella V, Ngiow SF, Khan O, Huang YJ, et al. (2020). Developmental Relationships of Four Exhausted CD8+ T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity 52, 825–841. 10.1016/j.immuni.2020.04.014. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

