Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression
- PMID: 35624211
- PMCID: PMC9174057
- DOI: 10.1038/s41590-022-01215-0
Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression
Abstract
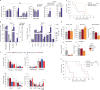
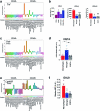
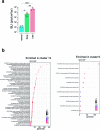
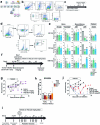
Glioblastoma (GBM) is an incurable primary malignant brain cancer hallmarked with a substantial protumorigenic immune component. Knowledge of the GBM immune microenvironment during tumor evolution and standard of care treatments is limited. Using single-cell transcriptomics and flow cytometry, we unveiled large-scale comprehensive longitudinal changes in immune cell composition throughout tumor progression in an epidermal growth factor receptor-driven genetic mouse GBM model. We identified subsets of proinflammatory microglia in developing GBMs and anti-inflammatory macrophages and protumorigenic myeloid-derived suppressors cells in end-stage tumors, an evolution that parallels breakdown of the blood-brain barrier and extensive growth of epidermal growth factor receptor+ GBM cells. A similar relationship was found between microglia and macrophages in patient biopsies of low-grade glioma and GBM. Temozolomide decreased the accumulation of myeloid-derived suppressor cells, whereas concomitant temozolomide irradiation increased intratumoral GranzymeB+ CD8+T cells but also increased CD4+ regulatory T cells. These results provide a comprehensive and unbiased immune cellular landscape and its evolutionary changes during GBM progression.
© 2022. The Author(s).
Conflict of interest statement
V.A.B. has patents on the PD-1 pathway licensed by Bristol-Myers Squibb, Roche, Merck, EMD-Serono, Boehringer Ingelheim, AstraZeneca, Novartis and Dako. The other authors declare no competing interests.
Figures













Comment in
-
Mapping the tumor-infiltrating immune cells during glioblastoma progression.Nat Immunol. 2022 Jun;23(6):826-828. doi: 10.1038/s41590-022-01223-0. Nat Immunol. 2022. PMID: 35624212 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

