Investigation and Comparison of Specific Antibodies' Affinity Interaction with SARS-CoV-2 Wild-Type, B.1.1.7, and B.1.351 Spike Protein by Total Internal Reflection Ellipsometry
- PMID: 35624652
- PMCID: PMC9139055
- DOI: 10.3390/bios12050351
Investigation and Comparison of Specific Antibodies' Affinity Interaction with SARS-CoV-2 Wild-Type, B.1.1.7, and B.1.351 Spike Protein by Total Internal Reflection Ellipsometry
Abstract
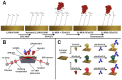
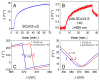
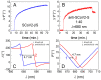
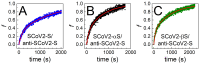
SARS-CoV-2 vaccines provide strong protection against COVID-19. However, the emergence of SARS-CoV-2 variants has raised concerns about the efficacy of vaccines. In this study, we investigated the interactions of specific polyclonal human antibodies (pAb-SCoV2-S) produced after vaccination with the Vaxzevria vaccine with the spike proteins of three SARS-CoV-2 variants of concern: wild-type, B.1.1.7, and B.1.351. Highly sensitive, label-free, and real-time monitoring of these interactions was accomplished using the total internal reflection ellipsometry method. Thermodynamic parameters such as association and dissociation rate constants, the stable immune complex formation rate constant (kr), the equilibrium association and dissociation (KD) constants and steric factors (Ps) were calculated using a two-step irreversible binding mathematical model. The results obtained show that the KD values for the specific antibody interactions with all three types of spike protein are in the same nanomolar range. The KD values for B.1.1.7 and B.1.351 suggest that the antibody produced after vaccination can successfully protect the population from the alpha (B.1.1.7) and beta (B.1.351) SARS-CoV-2 mutations. The steric factors (Ps) obtained for all three types of spike proteins showed a 100-fold lower requirement for the formation of an immune complex when compared with nucleocapsid protein.
Keywords: SARS-CoV-2; antibody–antigen interaction; immune complex; total internal reflection ellipsometry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- World Health Organization World Health Organization Tracking SARS-CoV-2 variants. [(accessed on 14 January 2022)]. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
-
- Global Virus Network Alpha (B.1.1.7)-GVN. [(accessed on 14 January 2022)]. Available online: https://gvn.org/covid-19/alpha-b-1-1-7/
-
- Bayarri-Olmos R., Johnsen L.B., Idorn M., Reinert L.S., Rosbjerg A., Vang S., Hansen C.B., Helgstrand C., Bjelke J.R., Bak-Thomsen T., et al. The alpha/b.1.1.7 sars-cov-2 variant exhibits significantly higher affinity for ace-2 and requires lower inoculation doses to cause disease in k18-hace2 mice. elife. 2021;10:1–14. doi: 10.7554/eLife.70002. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

